Extracellular vesicles (EVs), small membrane-bound structures secreted by various types of cells, are involved in cell-to-cell communication by transferring cargos such as nucleic acids [
12]. The delivery of nucleic acids, including microRNAs (miRNAs), through EVs to target cells may promote tumor growth and metastasis [
3]. The term ‘exosome’ is used to describe a population of small EVs (30–150 nm) that can be discriminated by size from microvesicles (100–1,000 nm) [
45]. However, the Minimal Information for Studies of Extracellular Vesicles 2018 study recommended the use of ‘small EVs’ rather than ‘exosomes’ for describing EVs sized <100 nm [
6]. For clinical utilization of small EVs in human disorders, it is essential to reproducibly isolate and enrich small EVs from blood and body fluids [
78].
Currently, several methodologies exist for the isolation and analysis of small EVs including ultracentrifugation (UC), size-exclusion chromatography (SEC), immunoaffinity capture-based methods, and polymer-based precipitation kits [
91011]. Although UC has long been considered the gold standard for isolating small EVs, there is an unmet need for methods that can completely separate small EVs from other nonvesicular entities such as apoptotic bodies.
Combinations of techniques have been evaluated in a bid to enhance the isolation efficiency and enrichment of small EVs. Recent studies have shown that coupling one cycle of UC with SEC provided better results than UC or SEC alone [
1213], although serum protein contamination remained a problem when using this approach [
13]. However, as studies utilizing human samples for clinical purposes usually require a large number of patients, polymer-based precipitation kits are used to isolate EVs from human samples because they do not require specialized equipment and have a large and scalable sample capacity. Despite this, polymer-based precipitation has limited value because it results in larger amounts of non-EV contaminants, such as proteins and polymeric materials, than other methods [
9]. To overcome this disadvantage, attempts have been made to use UC followed by enrichment using a polymer-based precipitation kit to isolate EVs from bovine milk; this combined method was reported to provide a better yield than polymer-based precipitation alone [
14]. However, there are limited data on the efficiency of coupling UC with a polymer-based precipitation technique for isolating EVs from the blood of cancer patients.
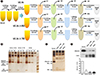
We used an approach combining UC and enrichment using one of the two commercially available polymer-based precipitation kits (ExoQuick [EQ], System Biosciences, Mountain View, CA, USA; or Total Exosome Isolation [TEI], Invitrogen Life Technologies, Carlsbad, CA, USA) to compare the isolation efficiency of UC alone (four cycles of UC [UC4x]) with that of UC followed by enrichment using EQ or TEI kits (two cycles of UC followed by the enrichment of small EVs using EQ [UC2x→EQ] or TEI [UC2x→TEI], respectively;
Fig. 1A). The isolated small EVs were characterized according to the recommendations for EV research and definition [
11].
Archived serum samples from breast cancer patients (N=30) that had been stored at −80℃ were used (Institutional Review Board of Samsung Medical Center, File No. SMC 2017-12-068, study period (2017–2019); the nine biosamples were provided by Samsung Medical Center BioBank, 2018-0004). As a preparation step, we first performed a brief round of low-speed centrifugation to eliminate cellular debris and large particles by sequentially centrifuging 1 mL of serum at 2,000×g at 4℃ for 10 minutes and 10,000×g at 4℃ for 30 minutes. The supernatant was then filtered through a 0.22-µm filter and ultracentrifuged at 110,000×g for 120 minutes at 4℃.
For UC4x, the samples underwent three further cycles of UC (110,000×
g) at 4℃ for 70 minutes, and the pellet was discarded at each step (
Fig. 1A). For UC2x→EQ and UC2x→TEI, the samples underwent two cycles of UC (110,000×
g) at 4℃ for 120 and 70 minutes, respectively. As all tests were conducted on the same sample, each group essentially comprised three samples containing 1 mL of serum. The resulting pellet was reconstituted in 0.5 mL of phosphate-buffered saline (PBS; Gibco, NY, USA) and added to 0.1 mL of EQ solution or TEI according to the manufacturer's protocols. The samples were incubated for 30 minutes at 4℃ and centrifuged twice at 1,500×
g for 30 and 5 minutes, respectively. The final pellet was reconstituted in 100 µL of PBS (
Fig. 1A).
The particle number of the isolated small EVs was quantified using the nanoparticle tracking analysis (NTA), and the protein concentration was assessed by the bicinchoninic acid (BCA) method using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Rockford, IL, USA). The small EVs were characterized by immunoblotting for the presence of EV markers (Western blotting) after the isolated EVs were lysed in RIPA buffer (0.5% sodium deoxycholate, 1% Nonidet P-40, 150 mM NaCl, 50 mM Tris [pH 7.5], 0.1% sodium dodecyl sulfate [SDS], and 1 mM phenylmethylsulfonyl fluoride). Ten micrograms of each protein sample was electrophoresed on a 4–12% SDS polyacrylamide gel and incubated overnight at 4℃ with the following antibodies: anti-CD63 (sc-5275), anti-CD81 (sc-7637), anti-Alix (sc-53540), anti-TSG101 (sc-7964) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and anti-albumin (Cell signaling Technologies, Danvers, MA, USA). The morphology of the isolated small EVs was analyzed by transmission electron microscopy (TEM).
Because the nucleic acid cargo carried by small EVs is important for their potential role in cancer patients, we extracted the RNA from the small EVs to evaluate the concentration of EV cargo miRNAs. As miR-21, miR-101, miR-155, miR-223, and miR-451a have been reported to be selectively enriched in EVs from patients with breast cancer, we measured their concentrations in small EVs [
15]. Small EV cargo miRNA was analyzed after total RNA was extracted from the isolated small EVs using the miRNeasy Micro Kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions. RNA concentration was measured using a NanoDrop ND-100 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The RNA was reverse-transcribed using the TaqMan microRNA Reverse Transcription Kit (Thermo Fisher Scientific) reagent containing specific miRNA primers (Thermo Fisher Scientific) for five miRNAs (miR-21-5p, miR-101-3p, miR-155-5p, miR-223-3p, and miR-451a), and real-time PCR (ABI PRISM 7900HT, Applied Biosystems, Foster City, USA) was performed in triplicate using the cDNA from each sample with 2× TaqMan Universal Master Mix II (with no AmpErase Uracil N-Glycosylase, UNG) and a 20× TaqMan miRNA expression assay (Thermo Fisher Scientific).
The differences between the three methods were analyzed by ANOVA using SPSS (version 23.0; IBM SPSS Inc., Armonk, NY, USA). P<0.05 was considered significant, and two-sided tests were used in all calculations. Graphs were plotted using GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA, USA).
Analysis of supernatants after the first round of UC revealed protein bands, including albumin and immunoglobulin heavy and light chains, which diminished after the final step of each method (EQ, TEI, and U4,
Fig. 1B). Silver-stained gels showed that the final pellets resuspended in PBS after the final step of the three methods (UC2x→EQ, UC2x→TEI, and UC4x) contained extremely low concentrations of albumin compared with the serum sample, although several bands reflecting contaminating proteins were still observed (
Fig. 1C). Western blotting for albumin and CD63 using 10 µg of protein from each sample also showed that the ratio of albumin to CD63 was significantly lower in the end-product of the three methods than in the serum sample (
Fig. 1D). These findings indicate that UC coupled to EQ or TEI might achieve an enrichment of small EVs comparable to that obtained using UC alone, as non-EV-related proteins, such as albumin and immunoglobulins, could be eliminated. The mean concentration of small EVs isolated from 1 mL of serum by UC2x→EQ and UC2x→TEI (139.0±29.1 µg and 140.4±5.0 µg, respectively) was not significantly different from that obtained using UC4x (141.8±26.9 µg,
Fig. 2A).
The concentration of small EVs we obtained was higher than that normally observed in other samples. This could be associated with the type of underlying disorders, as the concentration of small EVs isolated using UC4x from 1 mL of serum from healthy people and lymphoma patients in our previous pilot experiments was approximately 30 µg and 70 µg, respectively (data not shown).
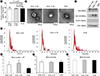
TEM imaging analysis showed that the small EVs isolated using the three methods were all rounded or cup-shaped and within the expected size range (
Fig. 2B). The small EVs isolated using the three methods were then characterized by Western blotting for CD63, CD81, Alix, and TSG101. Although CD63 expression was slightly lower in the small EVs isolated using UC2x→EQ, the expression of the other EV markers, including Alix, CD81, and TSG101, did not differ significantly between the three methods (
Fig. 2C).
The number and size of small EVs isolated by the three methods were analyzed using a Nanosight NS300 instrument (Malvern Instruments, Worcestershire, UK) with NTA (
Fig. 2D). The mean number of EV particles was 29.2±9.9×10
9 per mL of serum for UC4x, whereas UC2x→EQ and UC2x→TEI yielded a higher number of EV particles; however, there was no significant difference between UC2x→EQ (50.7±17.0×10
9 per mL of serum) and UC2x→TEI (59.3±20.6×10
9 per mL of serum,
Fig. 2E). The mean diameters (UC2x→EQ: 131.3±17.4 nm, UC2x→TEI: 127.4±11.8 nm, and UC4x: 130.5±23.1 nm) and mode diameters of the small EVs isolated using the three methods did not differ significantly (
Fig. 2F, G).
Taken together, these results indicate that 1 mL of serum could be the minimum volume required for our new method, similar to the UC method. As we had no internal control for EV miRNA, we extracted RNA from equal amounts of serum and determined miRNA concentrations. The concentrations of miR-21 and miR-155 in the small EVs isolated did not differ significantly among the three methods (
Fig. 3A, B); however, their concentrations differed from those in serum samples (
Fig. 3C). The difference between serum concentrations of miRNA and EV cargo miRNA might be associated with the different roles of circulating free miRNAs and exosomal miRNAs. The concentrations of miR-101, miR-223, and miR-451a also did not differ among the three methods (
Fig. 3D–F).
In conclusion, our results suggest that performing UC prior to using a polymer-based precipitation kit could be a feasible and convenient method for isolating small EVs from human serum in terms of yield and quality of EV cargos such as miRNAs. This approach may be useful for large sample-based translational researches, and further studies are warranted to validate its reliability for clinical application.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download