Abstract
Background
Differences in the performance of suggested warfarin dosing algorithms among different ethnicities and genotypes have been reported; this necessitates the development of an algorithm with enhanced performance for specific population groups. Previous warfarin dosing algorithms underestimated warfarin doses in VKORC1 1173C carriers. We aimed to develop and validate a new warfarin dosing algorithm for Korean patients with VKORC1 1173C.
Methods
A total of 109 patients carrying VKORC1 1173CT (N=105) or 1173CC (N=4) were included in this study. Multiple regression analysis was performed to deduce a new dosing algorithm. Following literature searches for genotype-guided warfarin dosing algorithms, 21 algorithms were selected and evaluated using the correlation coefficient (ρ) of actual dose and estimated dose, mean error, and root mean square error.
Results
The developed algorithm is as follows: maintenance dose (mg/week)=exp [3.223−0.009×(age)+0.577×(body surface area [BSA])+0.178×(sex)−0.481×(CYP2C9 genotype)+0.227×(VKORC1 genotype)]. Integrated variables explained 44% of the variance in the maintenance dose. The predicted and actual doses showed moderate correlation (ρ=0.641) with the best performance with a mean error of −1.30 mg/week. The proportion of underestimated groups was 17%, which was lower than with the other algorithms.
Warfarin is an oral anticoagulant used for the prevention and treatment of thromboembolic events [1]. Because of its narrow therapeutic index and significant interindividual variation in dose requirements, individualized dose adjustment is important for patient management. The most important factors affecting warfarin dose are VKORC1 and CYP2C9 polymorphisms, which explain approximately one-third of interindividual variations [23]. Pharmacogenetic-guided dosing decreases the time-to-stable doses and increases the percent time in therapeutic range compared with standard dosing [4].
Although these algorithms predict dosing requirements accurately in most patients, underestimations have been frequently observed in patients with higher dose requirements [567]. The meta-analysis conducted by Saffian, et al. [7] showed that all 22 algorithms included in the study underpredicted the warfarin dose for patients with a higher dose requirement. This can result in insufficient anticoagulation, leading to thrombotic events. In addition, because these algorithms show different performance depending on ethnicity, an ethnicity-specific algorithm is more appropriate for accurate dose prediction [8].
The VKORC1 1173C>T (complete linkage disequilibrium with −1639G>A, D′=1 and r2=1) is a common variant in the Asian population (82–96%) [1910]. Because of its association with warfarin sensitivity, patients with 1173CT and 1173CC genotypes require higher warfarin doses than patients with the 1173TT genotype [11]. Likewise, the high frequency of VKORC1 1173C>T in Asian populations can explain the lower warfarin dose requirements in Asians compared with those in Caucasians and Africans [9]. In our previous study, we evaluated the performance of 16 previously published warfarin dosing algorithms [6]. The 16 algorithms worked well for VKORC1 1173C non-carriers. However, they showed poor prediction performance in VKORC1 1173C carriers. An algorithm more suitable for VKORC1 1173C carriers is required for proper warfarin treatment and may be more important in populations with a high proportion of VKORC1 1173C carriers. Therefore, we developed and validated a new warfarin dosing algorithm for VKORC1 1173C carriers and compared its performance with that of previously published algorithms. This is the first study to develop and validate a warfarin dosing algorithm based on data from VKORC1 1173C carriers.
A total of 482 Korean patients with atrial fibrillation, cerebral infarction, or deep vein thrombosis/pulmonary embolism or patients undergoing valve replacement who were receiving warfarin treatment from 2006 to 2017 at the Samsung Medical Center, Seoul, Korea, were genotyped for CYP2C9 and VKORC1. After excluding 373 patients with the VKORC1 1173TT genotype, 109 patients with VKORC1 1173C were included. We retrospectively reviewed the clinical and laboratory data of these patients. Patient characteristics are presented in Table 1. High interindividual variation in actual dose was observed (median, 37 mg/week; range, 10–77 mg/week). For the VKORC1 genotypes, 96% of patients were heterozygous (VKORC1 1173CT) and 4% of patients were homozygous for the C allele (VKORC1 1173CC). For the CYP2C9 genotypes, 94% of patients had a *1/*1 genotype and 6% of patients had a *1/*3 genotype. All six patients with CYP2C9
*1/*3 had VKORC1 1173CT. None of the patients had CYP2C9
*1/*3 with VKORC1 1173CC. CYP2C9 and VKORC1 genotyping was performed using direct sequencing as described in our previous study [612]. This study was approved by the Institutional Review Board of Samsung Medical Center.
To develop the warfarin dosing algorithm, multiple linear regression analysis with backward variable selection was performed using the following clinical data and the genotyping results of the 109 patients: age, body surface area (BSA), sex, smoking, deep vein thrombosis, stroke, hypertension, diabetes mellitus, congestive heart failure, CYP2C9 genotype, and VKORC1 genotype. The distribution of warfarin dose was non-parametric, and warfarin dose was log-transformed for analysis. To evaluate the performance of our model, we used leave-one-out cross validation (LOOCV), which was deemed as the appropriate method because of the small study population. The i-th patient (i=1,…, 109) was divided from whole data as the test set and the remaining 108 patients as the training set [13]. Multiple linear regression analysis with backward variable selection was applied to the training set to fit a prediction model. A fitted prediction model was applied to the test set, and the predicted warfarin dose was calculated. This process was repeated for all patients. A scatter plot of the actual warfarin dose and predicted warfarin dose was generated after LOOCV, and its Spearman's correlation coefficient (ρ) was calculated. All statistical analyses were performed using SAS version 9.4 (SAS Institute, Cary, NC, USA).
For performance evaluation, the developed algorithm was compared with previously suggested ones. A total of 21 algorithms [41415161718192021222324252627282930313233], including CYP2C9 and VKORC1 genotypes, were selected through literature searches of PubMed for genotype-guided warfarin dosing algorithms published in the 2007–2017 period. We used a combination of search terms including warfarin, CYP2C9, VKORC1, dosing, dose prediction, algorithm, genotype-guided, and pharmacogenetic. Of the identified algorithms, we selected those published in English and those derived using the data of more than 100 patients. The 109 patients used for algorithm development were also included in the evaluation. The predictive performance of each algorithm was compared using the ρ of actual dose and estimated dose, mean error with 95% confidence interval (CI), and root mean square error. The actual maintenance dose was determined based on the weekly prescribed dose at three consecutive clinic visits when the international normalized ratio (INR) measurements were within the target range. The estimated dose was calculated for all tested algorithms using the incorporating factors of each algorithm. The value of ρ was interpreted as follows: negligible correlation (0–0.3), low correlation (0.3–0.5), moderate correlation (0.5–0.7), high correlation (0.7–0.9), or very high correlation (0.9–1.0) [34]. Mean error was calculated by subtracting the actual dose from the estimated dose. In addition, the percentages of patients with underestimated, ideal, and overestimated doses were evaluated. Patients with underestimated, ideal, and overestimated doses were defined as patients whose estimated dose was >20% lower than the actual dose, within 20% of the actual dose, and >20% higher than the actual dose, respectively.
Using multiple linear regression analysis, age, BSA, sex, CYP2C9 genotype, and VKORC1 genotype were selected for developing a warfarin dosing algorithm. The developed equation is as follows:
maintenance dose (mg/week)=exp[3.223−0.0094×(age)+0.577×(BSA)+0.178×(sex)−0.481×(CYP2C9 genotype)+0.227×(VKORC1 genotype)]
where exp is the exponential function.
In the case of females, the CYP2C9
*1/*3 genotype, or the VKORC1 1173CC genotype, it was coded as 1, while in the case of males, the CYP2C9
*1/*1 genotype, or the VKORC1 1173CT genotype, it was coded as 0. The developed algorithm explained 44% of the interindividual variations in the maintenance dose (ρ=0.641, P<0.0001). The LOOCV result is presented in Fig. 1. The ρ value between the actual and leave-one-out predicted dose was 0.664 (95% CI=0.543–0.757, P<0.001).
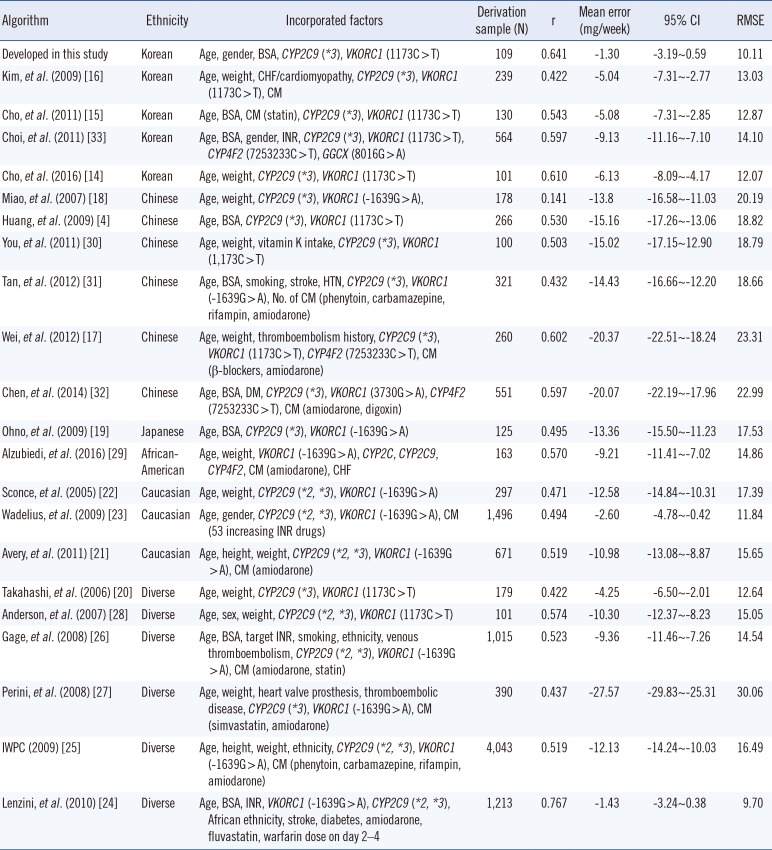
The ρ and mean error of the developed algorithm and the other 21 algorithms are presented in Table 2. The common factors included in all 22 algorithms were age, CYP2C9 genotype, and VKORC1 genotype. Ethnicity varied across the algorithms: five were derived from data of Koreans; six, from Chinese; one, from Japanese; three, from Caucasians; one, from African-Americans; and six, from diverse populations. The developed algorithm showed moderate correlation (ρ=0.641) and had the lowest mean error (−1.30 mg/week). The ρ and mean error of the other 21 algorithms ranged from 0.141 to 0.767 and from −1.43 to −20.37 mg/week, respectively. Of all 22 algorithms, the developed algorithm had the lowest proportion of underestimated groups (17%, Fig. 2). The algorithm by Lenzini, et al. [24] showed better correlation (ρ=0.767) than the developed algorithm; however, the performance, evaluated by the mean error and the proportion of underestimated groups was better for our algorithm.
Previously, we reported that warfarin dosing algorithms showed poor prediction performance in VKORC1 1173C carriers [6]. In that study, all 16 evaluated algorithms showed increased mean error in the VKORC1 1173C carrier group compared with that in the VKORC1 1173C noncarrier group. Here, we developed and validated a new warfarin dosing algorithm to predict maintenance doses for VKORC1 1173C carriers.
Previously, we developed a warfarin dosing algorithm with a derivation cohort that included 24.6% (32/130) VKORC1 1173C carriers [15]; by contrast, the present study included 100% VKORC1 1173C carriers. The derivation cohorts of the two studies were derived from the same hospital and showed very similar characteristics in terms of age, male:female ratio, body weight, and CYP2C9 genotype. Therefore, the different performance of the two algorithms could be caused by the different proportion of VKORC1 1173C carriers included in the derivation cohorts.
Considering the proportion of underestimated groups, the best-performing algorithm was the present algorithm, followed by those of Wadelius, et al. [23], Lenzini, et al. [24], and Takahashi, et al. [20]. The algorithms of Lenzini, et al. [24] and Wadelius, et al. [23] had the same proportion of underestimated groups (20%). The algorithm by Lenzini, et al. [24] had lower mean error, possibly due to the inclusion of Asian populations in the derivation cohort, and better correlation (ρ=0.767) compared with the present algorithm. However, the newly developed algorithm showed better performance, as estimated by mean error and the proportion of underestimated groups.
Although not the best, the algorithms derived from the data of Korean populations [14151633] showed relatively good performance even with low proportions of VKORC1 1173C carriers in the derivation cohort (Table 2; Fig. 2). In contrast, most of the algorithms derived from the data of different ethnic populations showed poor performance despite the high proportion of VKORC1 1173C carriers in the derivation cohort. These findings suggest that ethnicity is as important as VKORC1 genotype in predicting warfarin dose. Indeed, several studies have addressed the importance of developing ethnicity-specific warfarin dosing algorithms [568]. The current genotype-guided dosing algorithms were developed primarily based on the data from Caucasian populations and do not include genetic variants that are frequently found in non-Caucasian populations [2325]. The four algorithms derived from the data of Korean populations [14151633] were developed from derivation cohorts including 5.6–24.6% of VKORC1 1173C carriers. The proportion of VKORC1 1173C carriers was inevitably low in randomly selected derivation cohorts owing to the high frequency of VKORC1 1173C>T in the Asian population [1]. Because of the lower proportion of VKORC1 1173C carriers in the derivation cohort, these four algorithms showed a relatively higher mean error and higher proportion of underestimated groups than our algorithm. Therefore, the present algorithm developed for VKORC1 1173C carriers would be more helpful in Asian populations than in other ethnic populations.
Previous algorithms were developed mainly through multilinear regression analysis using clinical and genetic factors. A recent meta-analysis revealed that previous algorithms underestimated warfarin doses in patients requiring high doses, thus requiring the development of new algorithms for these patients [7]. To eliminate the bias effects caused by genetic factors, such as VKORC1 variants, developing ethnicity-specific algorithms similar to that by Limdi, et al. [8] could be a simpler approach. Therefore, the present algorithm, developed by regression analysis of clinical factors, is valuable because we targeted only specific genotype groups. We controlled the influence of VKORC1 1173C, which has significant effects on warfarin resistance in Koreans. However, these findings will require future validation in other ethnicities with VKORC1 1173C.
This study has several limitations. First, because of the small study population, we did not have an independent validation cohort. Therefore, performance evaluation was advantageous for our algorithm than for other algorithms developed using derivation cohorts with different characteristics. To overcome this limitation, we used LOOCV for performance evaluation. Further large-scale studies are required to validate our results. Second, we did not include the INR response in our analysis. According to Horne, et al. [35], incorporation of the INR response improves the performance of a warfarin dosing algorithm. However, as with our algorithm, most of the previous algorithms did not include the INR response. Third, our algorithm was not tested on other ethnic populations. Because ethnicity-specific algorithms are better than pan-ethnic algorithms, our algorithm may not work well in other ethnic populations. However, the primary goal of our study was to develop a more accurate algorithm for Korean VKORC1 1173C carriers. Fourth, although our algorithm had the lowest proportions of underestimated groups, 17% of patients were still underestimated. Therefore, our algorithm was not perfect and had a negative bias for some patients with higher dose requirements. Further studies are required to improve the performance of the algorithm for VKORC1 1173C carriers.
The algorithm developed in this study showed the best performance compared with 21 other algorithms. A warfarin dosing algorithm suitable for VKORC1 1173C carriers should be used to minimize the underestimation of warfarin dosing in these patients. It will enable proper treatment and reduce the risk of thromboembolism in VKORC1 1173C carriers.
Notes
AUTHOR CONTRIBUTIONS: Eun Hye Cho and Kyunghoon Lee performed data analysis and wrote the manuscript. Mina Yang, Rihwa Choi, June Soo Kim, Young Keun On, and Oh Young Bang collected data or contributed to data analysis. Sun-Young Baek and Insuk Sohn performed statistical analysis. Hyun-Jung Cho and Soo-Youn Lee designed the study, had full access to all of the data in the study, and take responsibility for the data integrity and the accuracy of the data analysis. All authors read and approved the final manuscript.
References
1. Shaw K, Amstutz U, Kim RB, Lesko LJ, Turgeon J, Michaud V, et al. Clinical practice recommendations on genetic testing of CYP2C9 and VKORC1 variants in warfarin therapy. Ther Drug Monit. 2015; 37:428–436. PMID: 26186657.
2. Kim S, Yun YM, Chae HJ, Cho HJ, Ji M, Kim IS, et al. Clinical pharmacogenetic testing and application: laboratory medicine clinical practice guidelines. Ann Lab Med. 2017; 37:180–193. PMID: 28029011.
3. Verhoef TI, Redekop WK, Daly AK, van Schie RM, de Boer A, Maitland-van der Zee AH. Pharmacogenetic-guided dosing of coumarin anticoagulants: algorithms for warfarin, acenocoumarol and phenprocoumon. Br J Clin Pharmacol. 2014; 77:626–641. PMID: 23919835.
4. Huang SW, Chen HS, Wang XQ, Huang L, Xu DL, Hu XJ, et al. Validation of VKORC1 and CYP2C9 genotypes on interindividual warfarin maintenance dose: a prospective study in Chinese patients. Pharmacogenet Genomics. 2009; 19:226–234. PMID: 19177029.
5. Peng Q, Huang S, Chen X, Yuan Y, Yu Y, Tao L, et al. Validation of warfarin pharmacogenetic algorithms in 586 Han Chinese patients. Pharmacogenomics. 2015; 16:1465–1474. PMID: 26255607.
6. Yang M, Choi R, Kim JS, On YK, Bang OY, Cho HJ, et al. Evaluation of 16 genotype-guided warfarin dosing algorithms in 310 Korean patients receiving warfarin treatment: poor prediction performance in VKORC1 1173C carriers. Clin Ther. 2016; 38:2666–2674.e1. PMID: 27887741.
7. Saffian SM, Duffull SB, Wright D. Warfarin dosing algorithms underpredict dose requirements in patients requiring ≥7 mg daily: a systematic review and meta-analysis. Clin Pharmacol Ther. 2017; 102:297–304. PMID: 28160278.
8. Limdi NA, Brown TM, Yan Q, Thigpen JL, Shendre A, Liu N, et al. Race influences warfarin dose changes associated with genetic factors. Blood. 2015; 126:539–545. PMID: 26024874.
9. Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005; 352:2285–2293. PMID: 15930419.
10. Patillon B, Luisi P, Blanché H, Patin E, Cann HM, Génin E, et al. Positive selection in the chromosome 16 VKORC1 genomic region has contributed to the variability of anticoagulant response in humans. PLoS One. 2012; 7:e53049. PMID: 23285254.
11. D'Andrea G, D'Ambrosio RL, Di Perna P, Chetta M, Santacroce R, Brancaccio V, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005; 105:645–649. PMID: 15358623.
12. Cho HJ, Sohn KH, Park HM, Lee KH, Choi B, Kim S, et al. Factors affecting the interindividual variability of warfarin dose requirement in adult Korean patients. Pharmacogenomics. 2007; 8:329–337. PMID: 17391071.
13. Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003; 95:14–18. PMID: 12509396.
14. Cho SM, Lee KY, Choi JR, Lee KA. Development and comparison of warfarin dosing algorithms in stroke patients. Yonsei Med J. 2016; 57:635–640. PMID: 26996562.
15. Cho HJ, On YK, Bang OY, Kim JW, Huh W, Ko JW, et al. Development and comparison of a warfarin-dosing algorithm for Korean patients with atrial fibrillation. Clin Ther. 2011; 33:1371–1380. PMID: 21981797.
16. Kim HS, Lee SS, Oh M, Jang YJ, Kim EY, Han IY, et al. Effect of CYP2C9 and VKORC1 genotypes on early-phase and steady-state warfarin dosing in Korean patients with mechanical heart valve replacement. Pharmacogenet Genomics. 2009; 19:103–112. PMID: 19077919.
17. Wei M, Ye F, Xie D, Zhu Y, Zhu J, Tao Y, et al. A new algorithm to predict warfarin dose from polymorphisms of CYP4F2, CYP2C9 and VKORC1 and clinical variables: derivation in Han Chinese patients with non valvular atrial fibrillation. Thromb Haemost. 2012; 107:1083–1091. PMID: 22534826.
18. Miao L, Yang J, Huang C, Shen Z. Contribution of age, body weight, and CYP2C9 and VKORC1 genotype to the anticoagulant response to warfarin: proposal for a new dosing regimen in Chinese patients. Eur J Clin Pharmacol. 2007; 63:1135–1141. PMID: 17899045.
19. Ohno M, Yamamoto A, Ono A, Miura G, Funamoto M, Takemoto Y, et al. Influence of clinical and genetic factors on warfarin dose requirements among Japanese patients. Eur J Clin Pharmacol. 2009; 65:1097–1103. PMID: 19582440.
20. Takahashi H, Wilkinson GR, Nutescu EA, Morita T, Ritchie MD, Scordo MG, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics. 2006; 16:101–110. PMID: 16424822.
21. Avery PJ, Jorgensen A, Hamberg AK, Wadelius M, Pirmohamed M, Kamali F, et al. A proposal for an individualized pharmacogenetics-based warfarin initiation dose regimen for patients commencing anticoagulation therapy. Clin Pharmacol Ther. 2011; 90:701–706. PMID: 22012312.
22. Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005; 106:2329–2333. PMID: 15947090.
23. Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, Bumpstead S, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009; 113:784–792. PMID: 18574025.
24. Lenzini P, Wadelius M, Kimmel S, Anderson JL, Jorgensen AL, Pirmohamed M, et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin Pharmacol Ther. 2010; 87:572–578. PMID: 20375999.
25. International Warfarin Pharmacogenetics Consortium. Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009; 360:753–764. PMID: 19228618.
26. Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008; 84:326–331. PMID: 18305455.
27. Perini JA, Struchiner CJ, Silva-Assunção E, Santana IS, Rangel F, Ojopi EB, et al. Pharmacogenetics of warfarin: development of a dosing algorithm for Brazilian patients. Clin Pharmacol Ther. 2008; 84:722–728. PMID: 18754001.
28. Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007; 116:2563–2570. PMID: 17989110.
29. Alzubiedi S, Saleh MI. Pharmacogenetic-guided warfarin dosing algorithm in African-Americans. J Cardiovasc Pharmacol. 2016; 67:86–92. PMID: 26355760.
30. You JH, Wong RS, Waye MM, Mu Y, Lim CK, Choi KC, et al. Warfarin dosing algorithm using clinical, demographic and pharmacogenetic data from Chinese patients. J Thromb Thrombolysis. 2011; 31:113–118. PMID: 20585834.
31. Tan SL, Li Z, Song GB, Liu LM, Zhang W, Peng J, et al. Development and comparison of a new personalized warfarin stable dose prediction algorithm in Chinese patients undergoing heart valve replacement. Pharmazie. 2012; 67:930–937. PMID: 23210243.
32. Chen J, Shao L, Gong L, Luo F, Wang J, Shi Y, et al. A pharmacogenetics-based warfarin maintenance dosing algorithm from Northern Chinese patients. PLoS One. 2014; 9:e105250. PMID: 25126975.
33. Choi JR, Kim JO, Kang DR, Yoon SA, Shin JY, Zhang X, et al. Proposal of pharmacogenetics-based warfarin dosing algorithm in Korean patients. J Hum Genet. 2011; 56:290–295. PMID: 21326313.
34. Mukaka MM. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012; 24:69–71. PMID: 23638278.
35. Horne BD, Lenzini PA, Wadelius M, Jorgensen AL, Kimmel SE, Ridker PM, et al. Pharmacogenetic warfarin dose refinements remain significantly influenced by genetic factors after one week of therapy. Thromb Haemost. 2012; 107:232–240. PMID: 22186998.
Fig. 1
Scatter plot showing Spearman correlation between actual and predicted warfarin doses using the leave-one-out cross validation (LOOCV) method (ρ=0.664, 95% CI: 0.543−0.757). The solid line is the regression line, and the dotted line is the 95% confidence interval.
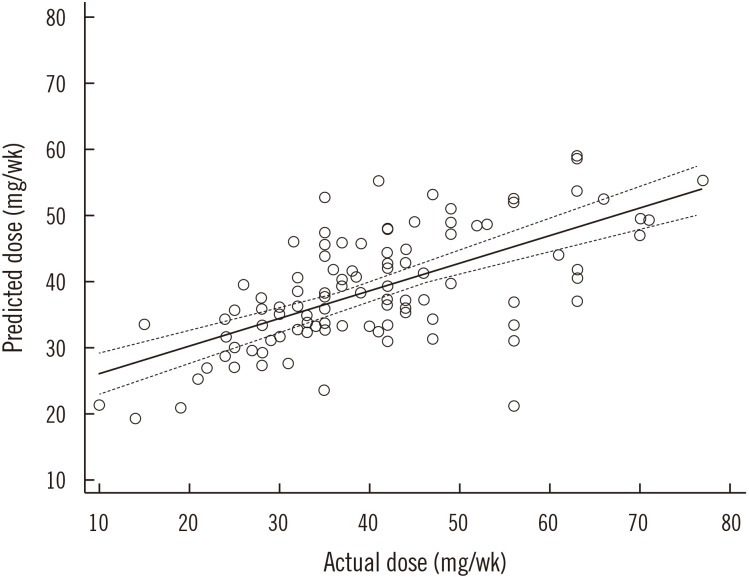
Fig. 2
Percentages of patients with underestimated (green), ideal (blue), and overestimated (red) doses of warfarin among VKORC1 1173C carriers.
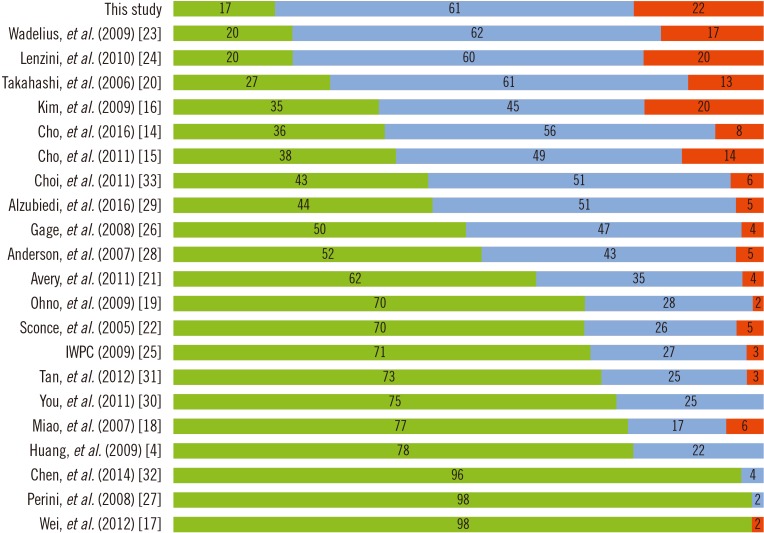
Table 1
Characteristics of the study population
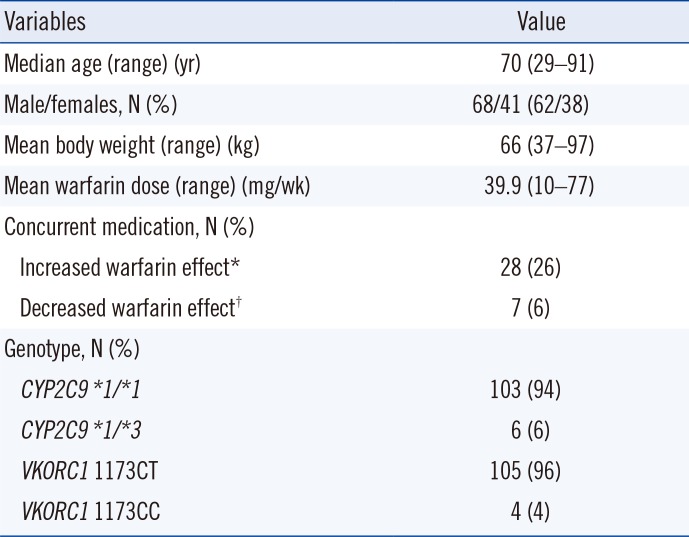
Table 2
Algorithms selected for performance evaluation in VKORC1 1173C carriers
| Algorithm | Ethnicity | Incorporated factors | Derivation sample (N) | r | Mean error (mg/week) | 95% CI | RMSE |
|---|---|---|---|---|---|---|---|
| Developed in this study | Korean | Age, gender, BSA, CYP2C9 (*3), VKORC1 (1173C > T) | 109 | 0.641 | -1.30 | -3.19~0.59 | 10.11 |
| Kim, et al. (2009) [16] | Korean | Age, weight, CHF/cardiomyopathy, CYP2C9 (*3), VKORC1 (1173C > T), CM | 239 | 0.422 | -5.04 | -7.31~-2.77 | 13.03 |
| Cho, et al. (2011) [15] | Korean | Age, BSA, CM (statin), CYP2C9 (*3), VKORC1 (1173C > T) | 130 | 0.543 | -5.08 | -7.31~-2.85 | 12.87 |
| Choi, et al. (2011) [33] | Korean | Age, BSA, gender, INR, CYP2C9 (*3), VKORC1 (1173C > T), CYP4F2 (7253233C > T), GGCX (8016G > A) | 564 | 0.597 | -9.13 | -11.16~-7.10 | 14.10 |
| Cho, et al. (2016) [14] | Korean | Age, weight, CYP2C9 (*3), VKORC1 (1173C > T) | 101 | 0.610 | -6.13 | -8.09~-4.17 | 12.07 |
| Miao, et al. (2007) [18] | Chinese | Age, weight, CYP2C9 (*3), VKORC1 (-1639G > A), | 178 | 0.141 | -13.8 | -16.58~-11.03 | 20.19 |
| Huang, et al. (2009) [4] | Chinese | Age, BSA, CYP2C9 (*3), VKORC1 (1173C > T) | 266 | 0.530 | -15.16 | -17.26~-13.06 | 18.82 |
| You, et al. (2011) [30] | Chinese | Age, weight, vitamin K intake, CYP2C9 (*3), VKORC1 (1,173C > T) | 100 | 0.503 | -15.02 | -17.15~12.90 | 18.79 |
| Tan, et al. (2012) [31] | Chinese | Age, BSA, smoking, stroke, HTN, CYP2C9 (*3), VKORC1 (-1639G > A), No. of CM (phenytoin, carbamazepine, rifampin, amiodarone) | 321 | 0.432 | -14.43 | -16.66~-12.20 | 18.66 |
| Wei, et al. (2012) [17] | Chinese | Age, weight, thromboembolism history, CYP2C9 (*3), VKORC1 (1173C > T), CYP4F2 (7253233C > T), CM (β-blockers, amiodarone) | 260 | 0.602 | -20.37 | -22.51~-18.24 | 23.31 |
| Chen, et al. (2014) [32] | Chinese | Age, BSA, DM, CYP2C9 (*3), VKORC1 (3730G > A), CYP4F2 (7253233C > T), CM (amiodarone, digoxin) | 551 | 0.597 | -20.07 | -22.19~-17.96 | 22.99 |
| Ohno, et al. (2009) [19] | Japanese | Age, BSA, CYP2C9 (*3), VKORC1 (-1639G > A) | 125 | 0.495 | -13.36 | -15.50~-11.23 | 17.53 |
| Alzubiedi, et al. (2016) [29] | African- American | Age, weight, VKORC1 (-1639G > A), CYP2C, CYP2C9, CYP4F2, CM (amiodarone), CHF | 163 | 0.570 | -9.21 | -11.41~-7.02 | 14.86 |
| Sconce, et al. (2005) [22] | Caucasian | Age, weight, CYP2C9 (*2, *3), VKORC1 (-1639G > A) | 297 | 0.471 | -12.58 | -14.84~-10.31 | 17.39 |
| Wadelius, et al. (2009) [23] | Caucasian | Age, gender, CYP2C9 (*2, *3), VKORC1 (-1639G > A), CM (53 increasing INR drugs) | 1,496 | 0.494 | -2.60 | -4.78~-0.42 | 11.84 |
| Avery, et al. (2011) [21] | Caucasian | Age, height, weight, CYP2C9 (*2, *3), VKORC1 (-1639G > A), CM (amiodarone) | 671 | 0.519 | -10.98 | -13.08~-8.87 | 15.65 |
| Takahashi, et al. (2006) [20] | Diverse | Age, weight, CYP2C9 (*3), VKORC1 (1173C > T) | 179 | 0.422 | -4.25 | -6.50~-2.01 | 12.64 |
| Anderson, et al. (2007) [28] | Diverse | Age, sex, weight, CYP2C9 (*2, *3), VKORC1 (1173C > T) | 101 | 0.574 | -10.30 | -12.37~-8.23 | 15.05 |
| Gage, et al. (2008) [26] | Diverse | Age, BSA, target INR, smoking, ethnicity, venous thromboembolism, CYP2C9 (*2, *3), VKORC1 (-1639G > A), CM (amiodarone, statin) | 1,015 | 0.523 | -9.36 | -11.46~-7.26 | 14.54 |
| Perini, et al. (2008) [27] | Diverse | Age, weight, heart valve prosthesis, thromboembolic disease, CYP2C9 (*3), VKORC1 (-1639G > A), CM (simvastatin, amiodarone) | 390 | 0.437 | -27.57 | -29.83~-25.31 | 30.06 |
| IWPC (2009) [25] | Diverse | Age, height, weight, ethnicity, CYP2C9 (*2, *3), VKORC1 (-1639G > A), CM (phenytoin, carbamazepine, rifampin, amiodarone) | 4,043 | 0.519 | -12.13 | -14.24~-10.03 | 16.49 |
| Lenzini, et al. (2010) [24] | Diverse | Age, BSA, INR, VKORC1 (-1639G > A), CYP2C9 (*2, *3), African ethnicity, stroke, diabetes, amiodarone, fluvastatin, warfarin dose on day 2–4 | 1,213 | 0.767 | -1.43 | -3.24~0.38 | 9.70 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download