1. Practice Committee of American Society for Reproductive Medicine. Definitions of infertility and recurrent pregnancy loss: a committee opinion. Fertil Steril. 2013; 99:63. PMID:
23095139.
2. Bareh GM, Jacoby E, Binkley P, Chang TC, Schenken RS, Robinson RD. Sperm deoxyribonucleic acid fragmentation assessment in normozoospermic male partners of couples with unexplained recurrent pregnancy loss: a prospective study. Fertil Steril. 2016; 105:329–336.e1. PMID:
26607021.

3. Rogenhofer N, Ott J, Pilatz A, Wolf J, Thaler CJ, Windischbauer L, et al. Unexplained recurrent miscarriages are associated with an aberrant sperm protamine mRNA content. Hum Reprod. 2017; 32:1574–1582. PMID:
28854581.

4. Dewan S, Puscheck EE, Coulam CB, Wilcox AJ, Jeyendran RS. Y-chromosome microdeletions and recurrent pregnancy loss. Fertil Steril. 2006; 85:441–445. PMID:
16595224.

5. Gil-Villa AM, Cardona-Maya W, Agarwal A, Sharma R, Cadavid A. Assessment of sperm factors possibly involved in early recurrent pregnancy loss. Fertil Steril. 2010; 94:1465–1472. PMID:
19540481.

6. Coughlan C, Clarke H, Cutting R, Saxton J, Waite S, Ledger W, et al. Sperm DNA fragmentation, recurrent implantation failure and recurrent miscarriage. Asian J Androl. 2015; 17:681–685. PMID:
25814156.

7. Gil-Villa AM, Cardona-Maya W, Agarwal A, Sharma R, Cadavid A. Role of male factor in early recurrent embryo loss: do antioxidants have any effect. Fertil Steril. 2009; 92:565–571. PMID:
18829003.

8. Ramasamy R, Scovell JM, Kovac JR, Cook PJ, Lamb DJ, Lipshultz LI. Fluorescence in situ hybridization detects increased sperm aneuploidy in men with recurrent pregnancy loss. Fertil Steril. 2015; 103:906–909.e1. PMID:
25707335.

9. Zidi-Jrah I, Hajlaoui A, Mougou-Zerelli S, Kammoun M, Meniaoui I, Sallem A, et al. Relationship between sperm aneuploidy, sperm DNA integrity, chromatin packaging, traditional semen parameters, and recurrent pregnancy loss. Fertil Steril. 2016; 105:58–64. PMID:
26493117.

10. Meyer RG, Ketchum CC, Meyer-Ficca ML. Heritable sperm chromatin epigenetics: a break to remember. Biol Reprod. 2017; 97:784–797. PMID:
29099948.

11. Mohanty G, Swain N, Goswami C, Kar S, Samanta L. Histone retention, protein carbonylation, and lipid peroxidation in spermatozoa: possible role in recurrent pregnancy loss. Syst Biol Reprod Med. 2016; 62:201–212. PMID:
26980262.

12. Simon L, Carrell DT. Sperm DNA damage measured by comet assay. Methods Mol Biol. 2013; 927:137–146. PMID:
22992910.

13. World Health Organization. WHO laboratory manual for the examination and processing of human semen. 5th ed. Geneva: World Health Organization;2010.
14. Baker MA, Witherdin R, Hetherington L, Cunningham-Smith K, Aitken RJ. Identification of post-translational modifications that occur during sperm maturation using difference in two-dimensional gel electrophoresis. Proteomics. 2005; 5:1003–1012. PMID:
15712234.

15. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951; 193:265–275. PMID:
14907713.

16. Hamada A, Sharma R, du Plessis SS, Willard B, Yadav SP, Sabanegh E, et al. Two-dimensional differential in-gel electrophoresis-based proteomics of male gametes in relation to oxidative stress. Fertil Steril. 2013; 99:1216–1226. PMID:
23312230.
17. Frapsauce C, Pionneau C, Bouley J, Delarouziere V, Berthaut I, Ravel C, et al. Proteomic identification of target proteins in normal but nonfertilizing sperm. Fertil Steril. 2014; 102:372–380. PMID:
24882558.

18. Samanta L, Agarwal A, Swain N, Sharma R, Gopalan B, Esteves SC, et al. Proteomic signatures of sperm mitochondria in varicocele: clinical use as biomarkers of varicocele associated infertility. J Urol. 2018; 200:414–422. PMID:
29530785.

19. Aoshima K, Inoue E, Sawa H, Okada Y. Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development. EMBO Rep. 2015; 16:803–812. PMID:
25925669.

20. Govin J, Caron C, Escoffier E, Ferro M, Kuhn L, Rousseaux S, et al. Post-meiotic shifts in HSPA2/HSP70.2 chaperone activity during mouse spermatogenesis. J Biol Chem. 2006; 281:37888–37892. PMID:
17035236.

21. Redgrove KA, Anderson AL, McLaughlin EA, O'Bryan MK, Aitken RJ, Nixon B. Investigation of the mechanisms by which the molecular chaperone HSPA2 regulates the expression of sperm surface receptors involved in human sperm-oocyte recognition. Mol Hum Reprod. 2013; 19:120–135. PMID:
23247813.

22. Bromfield EG, Aitken RJ, Anderson AL, McLaughlin EA, Nixon B. The impact of oxidative stress on chaperone-mediated human sperm-egg interaction. Hum Reprod. 2015; 30:2597–2613. PMID:
26345691.

23. Bironaite D, Pivoriunas A, Venalis A. Upregulation of iHsp70 by mild heat shock protects rabbit myogenic stem cells: involvement of JNK signalling and c-Jun. Cell Biol Int. 2012; 36:1089–1096. PMID:
22946646.

24. Jagadish N, Rana R, Selvi R, Mishra D, Garg M, Yadav S, et al. Characterization of a novel human sperm-associated antigen 9 (SPAG9) having structural homology with c-Jun N-terminal kinase-interacting protein. Biochem J. 2005; 389:73–82. PMID:
15693750.

25. Barroso G, Valdespin C, Vega E, Kershenovich R, Avila R, Avendaño C, et al. Developmental sperm contributions: fertilization and beyond. Fertil Steril. 2009; 92:835–848. PMID:
19631936.

26. Ishizuka M, Ohtsuka E, Inoue A, Odaka M, Ohshima H, Tamura N, et al. Abnormal spermatogenesis and male infertility in testicular zinc finger protein Zfp318-knockout mice. Dev Growth Differ. 2016; 58:600–608. PMID:
27385512.
27. Fan S, Zhao Y, Pan Z, Gao Z, Liang Z, Pan Z, et al. ZNF185-derived peptide induces fertility suppression in mice. J Pept Sci. 2018; 24:e3121. PMID:
30270484.

28. Meseguer M, de los Santos MJ, Simón C, Pellicer A, Remohí J, Garrido N. Effect of sperm glutathione peroxidases 1 and 4 on embryo asymmetry and blastocyst quality in oocyte donation cycles. Fertil Steril. 2006; 86:1376–1385. PMID:
16979635.

29. Puglisi R, Maccari I, Pipolo S, Mangia F, Boitani C. The nuclear form of glutathione peroxidase 4 colocalizes and directly interacts with protamines in the nuclear matrix during mouse sperm chromatin assembly. Spermatogenesis. 2014; 4:e28460. PMID:
25225625.

30. Ijiri TW, Merdiushev T, Cao W, Gerton GL. Identification and validation of mouse sperm proteins correlated with epididymal maturation. Proteomics. 2011; 11:4047–4062. PMID:
21805633.

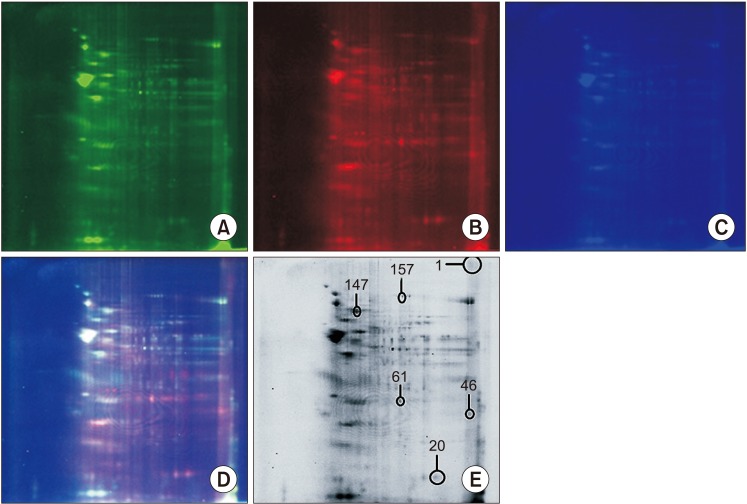

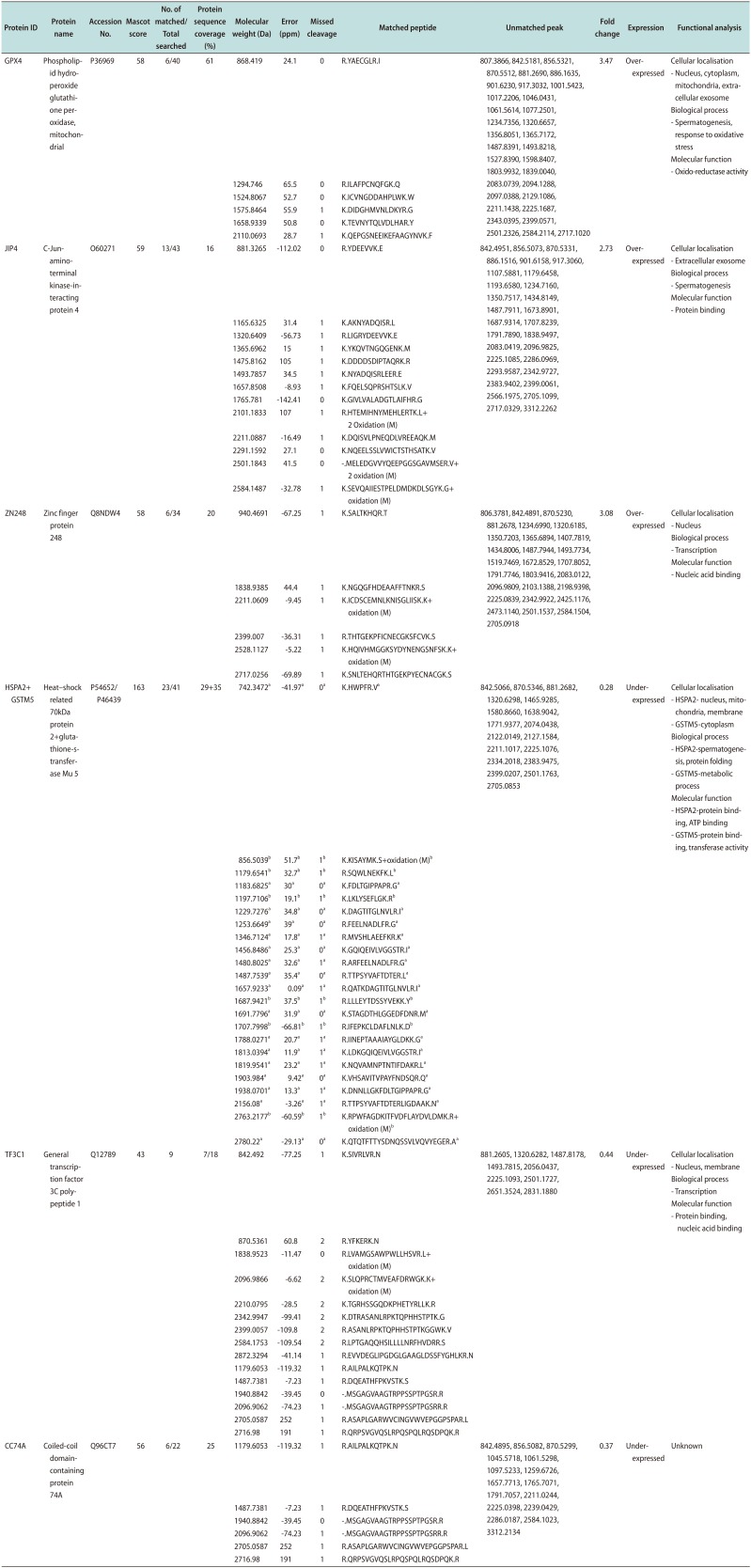
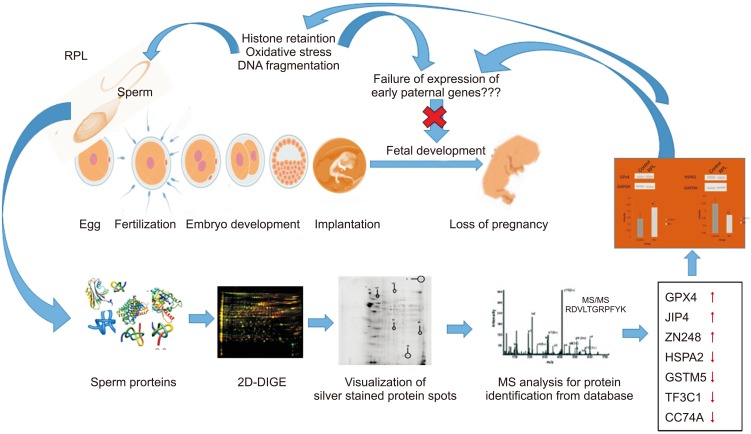




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download