Abstract
Microtubules are the prime component of the cytoskeleton along with microfilaments. Being vital for organelle transport and cellular divisions during spermatogenesis and sperm motility process, microtubules ascertain functional capacity of sperm. Also, microtubule based structures such as axoneme and manchette are crucial for sperm head and tail formation. This review (a) presents a concise, yet detailed structural overview of the microtubules, (b) analyses the role of microtubule structures in various male reproductive functions, and (c) presents the association of microtubular dysfunctions with male infertility. Considering the immense importance of microtubule structures in the formation and maintenance of physiological functions of sperm cells, this review serves as a scientific trigger in stimulating further male infertility research in this direction.
Infertility is the inability of a couple to achieve pregnancy after at least one year of unprotected sexual intercourse and male infertility accounts for about half of the total number of infertility cases [1]. Male infertility is a heterogeneous and multifactorial condition with several etiologies including varicocele, antisperm antibodies, undescended testis, hypogonadism and a whole range of genetic disorders, such as aneuploidies and structural chromosomal aberrations and affects more than 30 million males worldwide [2].
Sperm with normal morphology and motility is essential for normal male fertility [3]. Microtubules and microfilaments play pivotal roles in maintenance of these sperm qualities during different phases of sperm production and maturation [45]. In dividing germ cells, bipolar spindle, a complex and dynamic microtubule assembly, is involved in the segregation of chromosomes and positioning of the cell-division plane [6]. In spermatogenic cells, microtubules are necessary for several of processes, including the assembly of flagella in spermatids [45], and the generation and maintenance of motility of mature spermatozoa [5].
Spermatozoa are the smallest and most polarized cells in the body with a unique structure made up of a head not only containing the genetic material, but also all the enzymes in the acrosome necessary for the penetration of the oocyte vestments (cumulus and zona pellucida) in order to fertilize the egg. On the other hand, there is the sperm tail or flagellum comprising the mid (connecting piece), principal and end piece [7].
Microtubles are the central element of cilia and flagella. Since they play key roles in human reproduction, any defects in their structure can lead to infertility. In this review, we aimed to discuss the structure of microtubules and the role they play in male reproduction and male infertility. We also provide an overview of the known genes associated with microtubular formation and function, particularly the axoneme and manchette on various sperm phenotypes of genetic origin.
Microtubules are composed of a globular alpha (α)- and beta (β)-tubulins, which polymerize using guanosine-5′-triphosphate (GTP) to form a single protofilament (Fig. 1) [8]. The tubulins are arranged in a polarized manner whereby the α-tubulins are exposed at the negative (−) and the β-tubulins at the positive (+) end. The dynamic properties of the two microtubules ends are different. The (−) end is more stable and grows slowly whereas the (+) end can polymerize (grow) and depolymerize (shrink) rapidly, thus rendering microtubules highly dynamic structures [9]. The GTP bound to β-tubulin hydrolyzes during polymerization, but the GTP attached to α-tubulin neither hydrolyze nor exchange and remains as an integral part of the αβ heterodimer in the microtubule. The conversion of GTP to guanosine 5′-diphosphate in β-tubulins via hydrolysis dictates whether the microtubule will grow or shorten (Fig. 1) [8].
In vivo, αβ-heterodimers require a nucleator such as γ-tubulin so that microtubules can start grow and orientate properly at the poles. γ-tubulins are found in complexes known as γ-tubulin ring complexes (γTuRC) that function as a nucleation sites. The γTuRC chemically mimic the (+) end of a microtubule and thus allow microtubules to bind [9].
Microtubules and actins are the main building blocks of the cytoskeleton along with intermediate filaments. These hollow tubes are nucleated from the center of an interphase cell and form bipolar mitotic spindles during cell division. They support the shape of the cell, organize the intracellular structures, and aid in intracellular transport [1011].
In the developing spermatid, the centrosome is the only distinct microtubule organizing center (MTOC) that consists of two L-shaped centrioles, a parent and a daughter centriole, surrounded by the peri-centriolar matrix (in developing spermatozoa the other important MTOC are the basal bodies which are concerned with cilia formation). In dividing cells, the centrosome divides once per each cell cycle [12], and become the basal bodies of the cilia and flagella in motile cells like spermatozoa.
The manchette is a transient horizontal structure surrounding the elongating spermatid head and is present only during spermatid elongation [13]. This temporary structure is responsible for sperm tail formation and head shaping during spermatogenesis [14] and then disappears in mature sperm [15]. The presence of the manchette can first be noticed at the beginning of the elongation phase of spermiogenesis and disassembles prior to the mid-piece of sperm tail formation [9].
The manchette creates a platform consisting of microtubules and actin filaments between the perinuclear ring and the elongated sperm axoneme [16]. It also elongates the spermatid head as it is associated with the nuclear shaping [17]. The perinuclear ring and centrosome play a part in connecting and stabilizing the manchette's microtubules after they are formed [18]. The manchette is composed of microtubules and fibrous (F)-actin filaments. The F-actin filaments and actin-dependent molecular motor protein, myosin play fundamental roles in the transport of vesicles along the F-actin and in the shaping the spermatid head. Nuclear reconstruction during sperm head formation coincides with modulation of the nuclear envelope, which in turn is presumed to be critical for transferring cytoskeletal forces, and required for directed nuclear shaping [19]. The manchette seems to be connected through linker of nucleoskeleton and cytoskeleton (LINC) complexes. LINC complexes are identified as nuclear envelop-bridging assemblies connecting the nuclear content to the cytoskeleton. These bridges are formed by interaction of two evolutionarily conserved transmembrane protein families: the SUN (Sad-1/UNC-84) and KASH (Klarsicht/ANC-1/Syne/homology) domain proteins [20]. Two LINC complexes, SUN1/Nesprin3 and SUN3/Nesprin1, locate at opposite poles during spermatid elongation [21]. SUN3/Nesprin1 complex links the manchette to the nuclear envelope and SUN1/Nesprin3 may be involved in basal body attachment to the nucleus. SUN proteins (with testis specific SUN3, 4, and 5) represent the inner nuclear membrane constituents of LINC complexes [19]. Calvi et al [22] found that loss of SUN4, a spermatid nuclear membrane protein, leads to disorganization of the microtubules and a failure of the nucleus to elongate. Moreover, SUN4-deficient mice found to have globozoospermia with associated infertility.
KASH proteins reside within the outer nuclear membrane and connect to the cytoskeleton [23]. These proteins, along with the LINC complex, provide physical force to shape the sperm head by constructing the nuclear DNA to form the distal half of the nucleus.
Intra-manchette transport (IMT) and intra-flagellar transport (IFT) are two processes that take place during spermiogenesis and are crucial for male fertility. Both IMT and IFT are absent in mature sperm [17]. IMT transfers structural and functional proteins that form the sperm tail via the microtubule tracks and motor proteins to the basal body region where they are stored. Moreover, IMT is also responsible for nucleocytoplasmic transport. The depletion of dynein-related proteins (CLIP-170) leads to abnormal nucleus shape [18].
IFT is responsible for sperm-protein transportation during the development of the flagella [2425]. During IFT, cargos are transported from the sperm cytoplasm to the tip of the flagellum and then back to the sperm head along the axonemal microtubules. Outward or anterograde movement (from the sperm head to the tail) is directly connected with microtubule motor kinesin-2 while inward or retrograde movement is linked up cytoplasmic dynein-2 (dynein 1b) [18]. Although the precise mechanisms by which the specific cargos are stored and transported during IMT and IFT are not known, microtubules, which are arranged in opposite directions in these two processes, along with dynein, a candidate motor protein for the LINC complex, play crucial roles for spermatid head modelling during spermiogenesis [26]. Mutations in genes associated with the IMT or IFT microtubule structures are responsible sperm head and tail abnormalities because they can disrupt protein delivery to the correct assembly site during spermiogenesis and thus lead to abnormalities in mature sperm (Fig. 2) [27].
The flagellum starts growing from the basal body (i.e., the daughter centriole of the centrosome) while this structure migrates towards the cell membrane. Afterwards, the centrosome-axoneme complex migrates and attaches to the nucleus. Affixed and stabilized by the basal body, acting as MTOC, the microtubules and thereby the flagellum grow in a directed manner. Flagellar ultrastructure is divided into four main parts: the connecting piece, the mid piece, the principal piece, and the end piece. The connecting piece consists of distal and proximal centrioles. The mid piece contains a ring-shaped mitochondrial sheath surrounding the axoneme to provide the energy for the movement of flagella. Principal piece contains fibrous sheath around the axoneme. The last part, end piece, incorporates only the axoneme [14].
Apart from mitochondria located in the mid piece, the outer dense fibers and the fibrous sheath, the flagellum contains the microtubular structures the axoneme and the manchette, which play important roles during part of the formation of sperm head and tail [18]. Defects of the microtubules or microtubule-related components may cause sperm abnormalities and infertility problems such as teratozoospermia, oligozoospermia, asthenozoospermia and even azoospermia [28].
The axoneme is an essential structure for the generation of sperm motility. This structure originates from the distal centriole of the round spermatid centrosome and then comes into contact with the nuclear membrane on the opposite pole of the acrosome [9]. Axoneme is the inner core structure of cilia and flagella, and is set up by microtubules in a typical 9+2 pattern with one central pair (CP) and nine peripheral microtubules doublets. Regulation and modulation of ciliary movement is controlled by CP, inner dynein arms (IDAs), and nexin-dynein regulatory complex, however beat generation is controlled by the outer dynein arms (ODAs) [29]. The axoneme also possesses radial spokes that connect the central and peripheral microtubules and are also related to the mechanical movement of the flagellum (Fig. 3). Consecutive microtubules are linked by nexin, a protein that stabilizes the axoneme [14]. Axoneme structures play an essential role in the morphology and functioning of the flagellum. Any alteration in the axoneme ultrastructure may cause abnormalities in the sperm tail and change its morphology, causing severe motility disorders [30].
Four types of microtubule-associated proteins (MAPs) interact with tubulin in specific ways to regulate microtubule dynamics and organization, namely: motor proteins, crosslinking proteins, microtubule number regulators, and dynamic instability regulators [31].
Motor proteins are adenosine triphosphate (ATP)-dependent molecular motors that transport cargo along the microtubule and deliver the contents to its designated location within the cell (e.g., kinesins and dynein) [32]. Microtubule plus-end-tracking proteins (+TIPs) are evolutionarily conserved motor proteins that bind to microtubule plus (+) ends when polymerization is initiated and play a crucial role in microtubule dynamics. Microtubule +TIPs function as microtubule-stabilizing factors and can alter plus (+) end dynamics at microtubule-kinetochore binding sites in dividing cells [33]. Crosslinking proteins help to stabilize the structure of cellular components [34] and microtubule number regulators control nucleation mechanism [35], whereas dynamic instability regulators control the co-existence of microtubular assembly and disassembly [36]. Different combinations of these four types of MAPs work together to assemble microtubule-based structures in various cellular processes [31].
Microtubules play important roles in male fertility and ensure successful cell division during spermatogenesis. Fawcett [13] in his pioneering article on mammalian spermatozoa has discussed the role of microtubular structures in spermatogenesis. Bertalan et al [37] found that the total number of microtubules in the mitotic spindle is a critical controlling factor in cell division. If the number of microtubules significantly changes, this may result in altered cell division with subsequent pathological developments. Microtubule polymers build the mitotic spindle, and numerous associated factors unite to create a dynamic steady state structure [38]. Mitotic spindles in spermatogonia orient perpendicularly to the basement membrane to aid mitotic cell division and are rare in adults, suggesting a faster rate of mitotic division [39].
Spermatogonia, proliferative type A, undergo a series of mitotic divisions to reproduce themselves. While one of these daughter cells restores of type A spermatogonial stock, the other changes to type B spermatogonia in the basal compartment. Then, type B spermatogonia differentiate to form pre-leptotene spermatocytes, which are transported across the seminiferous tubule barrier [40]. After committing to meiosis, two cellular divisions occur where the 4N primary spermatocyte divides to form two 2N (N refers to haploid genome) secondary spermatocytes. Subsequently, each secondary spermatocyte divides to form two N spermatids [41]. The segregation of chromosomes involves three types of microtubules: kinetochore, astral, and polar. The kinetochore microtubules bind to the kinetochore at the chromosomal centromere which is critical for proper chromosomal segregation [38]. Errors in chromosome segregation lead to aneuploidy due to nondisjunction in both mitosis and meiosis. Astral microtubules are cell membrane interacting microtubules and are not connected directly to the chromosome. However, they participate in spindle positioning and cleavage before cytokinesis. Polar microtubules interlace at the midzone of the spindle and aid propelling of the spindle pores apart by the motor proteins [9].
During spermiogenesis, spermatids differentiate into spermatozoa, a process which involves four phases: the Golgi phase, the cap phase, formation of the tail, and the maturation phase. This process was classically described by Holstein [42] using ultrastructural investigations that confirmed the role of microtubules and axonemal structure in spermatid maturation. The Golgi stage of spermiogenesis is characterized by the formation of the head and axoneme. During this stage, sperm head shaping begins with the polarization of round spermatid nucleus to the one side of the cell and then proceed to deviate from a spherical shape as nuclear condensation continue. In addition, the acrosomal cap is formed over the anterior surface of the nucleus [9]. Normal sperm nuclear morphology relies on sperm DNA compaction as well as specialized structures within spermatids that determine the characteristic species-specific sperm head shape. As discussed earlier, the two major structures associated with sperm head shaping, known as the acroplaxome and manchette are closely related to one another as spermatids proceed through the elongation phase [43]. Later in this phase, about 90% of nuclear histones are removed and spermatozoal DNA is packaged first with specific nuclear proteins called transition proteins. Then, these transition proteins are replaced with specialized proteins called protamines [9]. As the acrosome begins to form on one pole of the nucleus, the two pairs of centrioles move toward the opposite pole to initiate axoneme formation. During formation of the sperm tail, the basal body (which is assembled within the spermatid centrosome) attaches with the microtubules. Microtubules present in this region are responsible for trafficking vesicles from the Golgi apparatus to the acrosome [44]. After the initiation of axoneme formation in early spermiogenesis, secondary structures necessary for flagella function (outer dense fibers, the fibrous sheath and the mitochondrial sheath) are assembled during the elongation phase of spermiogenesis [45]. Followed by the formation of the sperm tail, mature spermatozoa get released from Sertoli cells into the lumen of the seminiferous tubule in a process called spermiation [9]. During spermiation, residual bodies are phagocytized by the Sertoli cells [46]. This process involves removal of specialized adhesion structures and remodeling of the spermatid head and cytoplasm [9].
Several proteins and their interactions have been reported to play significant roles during sperm development. For example, Hook1 is a protein that binds to microtubules and cargoes in Drosophila [47]. This protein belongs to the family of Hook proteins, consisting of Hook1, Hook2, and Hook3 in mammals. Hook 2 has been reported to play a role in primary cilia morphogenesis while Hook3 has been shown to participate in the localization of Golgi complex [48]. A recent study described the interaction of Hook1 with another protein called coiled-coil domain containing protein 181 (Ccdc181) during localization of the sperm tail and in the formation of motile cilia [49]. Hook1 gene deletion has been reported to cause disruption of the manchette structure and abnormal head morphology in mice. Hook1 gene is suggested as a candidate for decapitation defects and teratozoospermia [50]. The different steps of spermatogenesis and microtubule functions are summarized in Fig. 4.
The tail is a long flagellum, and the central axoneme of this flagellum emerges from a basal body located posterior to the nucleus. For sperm motility, sliding of outer doublet microtubules in the sperm flagellum needs energy generated by ATP hydrolysis [11]. In human sperm, ATP is produced by glycolysis in the principle piece of the flagellum [51] and also, by the mitochondria present in the mid piece [52].
Katanin is a microtubule-severing complex consisting of a p60 severing enzyme and a p80 regulatory subunit [53]. Katanin p80 plays a crucial role in meiotic spindle formation, polarization and depolarization of microtubules and reproduction in male mice [54]. The protein p80 is encoded by Katnb1, and the missense mutation of this gene causes male infertility in mice characterized by oligoasthenoteratozoospermia. Mutation of this gene also causes virtual absence of progressive motility [54]. The expression of katanin p80 (both mRNA and protein levels) was evaluated in testis biopsies from patients with maturation arrest at the level of spermatocyte and spermatogonia and with Sertoli cell only syndrome, and compared with samples showing normal spermatogenesis [55]. These results suggest that KATNB1 plays a role in meiosis, nuclear shaping, and flagellum formation of sperm in humans. It was also reported that expression of p80 occurred only in germ cells and not in stem cells, Sertoli cells or interstitial cells [54].
In mammals, the blood-testis barrier (BTB) creates a unique microenvironment for the development and maturation of germ cells in order to maintain and support spermatogenesis. The BTB is located between Sertoli cells and germ cells, as well as between germ cells [56]. Sertoli cells are dynamic, metabolic epithelial cells that support the seminiferous epithelium. These cells contain microtubules that are oriented in linear arrays parallel to the long axis of the cell, forming a cage-like structure around the nucleus [57]. The shape of Sertoli cell changes according to the requirement of the developing germ cells [9].
The Katanin catalytic subunit A1 like 1 (KATNAL1) gene is a protein coding gene that is expressed in testicular tissue and helps control Sertoli cell microtubule dynamics [58]. Loss of function mutations of KATNAL1 results in disruption of Sertoli cell microtubule dynamics and cause to release of immature sperm within the testis tubules during sperm maturation [59]. Furthermore, it has been reported that induced-expression of a functional KATNAL1 protein raises microtubule severing, thereby impairs Sertoli cell plasticity required for germ cell growth. In the same study, it has also been shown that co-localization of with a Sertoli cell-specific isoform β-tubulin ‘TUBB3’ in Katnal11H/1H mutant testes [60]. Localization of TUBB3 showed that microtubule network underwent enormous disruption in the mutant testis [59]. Accordingly, a hypothesis was made that over-expression of functional KATNAL1 disrupts microtubule dynamics, which results in cellular arrest during mitosis that eventually leads to cell death [61]. However, the proper localization, mechanism of nucleation in Sertoli cell microtubules and its control are still not clear.
A-kinase anchor proteins (AKAPs) are regulators of microtubule dynamics and contribute to normal BTB function. Akap9 deletion has been reported to lead to significant changes in the microtubule organization in Sertoli cells and loss of BTB in Akap9 null mice [56].
Male infertility caused by impaired sperm motility and defects in genes that encode for axonemal complexes can be observed in men with primary ciliary dyskinesia (PCD) [62]. It was first described as a syndrome based on the triad of chronic sinusitis, bronchiectasis and situs inversus [63]. Forty years later, in the mid-1970s, Afzelius [64] expanded on this syndrome by observing that these patients had ‘immotile’ cilia and defective ciliary ultrastructure, specifically noting a deficiency of dynein arms, decreased mucociliary clearance, and a lack of ciliary motion. Presently, it is well-known as a heterogeneous and multisystemic disorder that is characterized by structural or functional abnormalities of the motile cilia [65].
PCD is a rare autosomal recessive disorder with an estimated prevalence of 1:10,000 to 1:20,000 in the general population [66]. In 1975, Afzelius et al [67] first reported PCD as a genetic disorder involving production, assembly, or attachment of dynein arm in spermatozoa in two brothers who were not twins. Pedersen and Rebbe [68] reported total immobility of spermatozoa in a patient with absent dynein arms in the axoneme of spermatozoa. Approximately, one half of patients presenting with PCD have situs inversus, a condition where the major organs appear mirrored to their normal position, which is called Kartagener syndrome [69]. Males with PCD are infertile due to immotile sperm and are at an increased risk of having respiratory problems such as lung infections caused by paralysis of the cilia [70]. Nearly 30 genes are involved in PCD pathogenesis in humans [71].
Various defects in the cilia structure are present in patients with PCD, including radial spoke abnormalities, outer and/or IDAs defects and the absence of a central microtubule [30]. Morphological changes always accompany one of these defects [72]. Abnormalities of the cilia can be classified as (1) dynein arm abnormalities with the most common cilia abnormality [73] involving either partial or complete absence of ODA/IDA [74]; (2) radial spoke abnormalities, including the absence of spokes or deviated central microtubules [75]; (3) inappropriate directionality of cilia, caused by a partial or complete absence of the central microtubules or defects in planar cell polarity [74]; and (4) an abnormal number of peripheral microtubules such as, 8+1, 8+2, 8+3, or 7+2 arrangements of the cross-section (Fig. 5) [76].
PCD occurs as a result of genetic mutations in genes encoding for ciliary proteins [77]. Many structural and regulatory proteins are present in the cilia, and hundreds of genes control these proteins. As a result, mutation in any of the genes encoding for these proteins may lead to abnormalities in ciliary functions [78].
Several studies have focused on identifying genes responsible for PCD [2979] using genetic linkage analysis in familial cases and candidate gene approaches, and proteomic analyses [80]. The most commonly studied genes are the dynein axonemal intermediate chain 1 (DNAI1) and dynein axonemal heavy chain 5 (DNAH5) genes [2980]. Mutations in these two genes result in abnormalities in ciliary structure and motor functions [2980]. DNAI2 is expressed in the trachea and testes and its mutations can lead to PCD [81]. Mutations in dynein axonemal beta heavy chain 9 (DNAH9) and dynein axonemal heavy chain type 11 (DNAH11) were found to be involved in PCD [79]. Mazor et al [82] reported a homozygous point mutation of dynein axonemal light chain 1 (DNAL1) that reduced the axonemal dynein light chain 1 stability and damaged its interactions with tubulin and with dynein heavy chain, subsequently resulting in PCD. Three additional mutations have been identified in DNAAF1, DNAAF2, and DNAAF3 all of which play important roles in the preassembly of dynein into complexes before their transport into cilia [83].
Castleman et al [84] reported two novel mutations in radial spoke head 9 homolog (RSHP9) and radial spoke head 4 homolog A (RSPH4A) encoding radial spoke head proteins with defects of the central microtubular pair related with PCD. These authors performed homozygosity mapping and identified the location of these genes on the long arm of chromosome 6. This study also revealed that RSPH9 is important in maintaining normal movement of motile ‘9++2’ structure of flagella. Ultrastructure analysis showed transposition defects with the absence of CP and 9+0 or 8+1 microtubule configuration.
Most genes implicated in PCD were thought to cause structural defects in the axoneme through mutations inducing CP defects. Recently, an association between a homozygous loss-of-function mutation in DNAJB13, a heat-shock protein, and PCD has been shown. DNAJB13 plays an essential role in preserving the integrity of the CP in humans [85]. Gene mutations and their consequences for axonemal structure in human and functional defects in PCD are summarized in (Table 1).
Dysplasia of the fibrous sheath (DFS) is an autosomal recessive genetic condition and appears in severe asthenozoospermic patients, which are characterized by dysplastic development of the axonemal and peri-axonemal cytoskeleton [86]. DFS is diagnosed by marked hypertrophy and hyperplasia of the fibrous sheath in association with deficiency of the annulus, mitochondria, the CP complex (50% of the cases) and/or the dynein arms [86]. The genetic origin of this disorder has been reported by numerous researchers [8788].
The AKAPs, which are the most abundant structural proteins of the fibrous sheath and engaged in organizing and assembling the basic structure of protein (AKAP3, 4) [89] have been reported to cause DFS on its mutation [88]. In humans, two other forms of these sperm fibrous sheath proteins, hAKAP82 and its precursor pro-hAKAP82, were cloned and characterized [90]. DFS is prevalent in consanguineous marriages [87] and is associated with PCD (in 20% of the cases), transmission of unbalanced gametes [8891], and increased sperm ubiquitination [92].
Several studies have reported a correlation between sperm flagellar abnormalities and increased frequency of gonosomal disomies and diploidies in spermatozoa [93]. In addition, numerous studies have found an elevated aneuploidy rate in multiple morphological abnormalities of the sperm flagella (MMAF), which might be associated with the sperm centrosome, the mitotic spindle and the flagella [94]. An elevated aneuploidy rate suggests that abnormalities of various centrosome-associated proteins may affect flagellum formation and mitotic/meiotic spindle assembly during spermatogenesis and lead to numerical chromosomal aberrations in spermatozoa [93]. However, these chromosomal aneuploidies have not been observed in all MMAF patients, suggesting that these flagellar and spindle defects may not directly cause chromosomal aneuploidies in all cases [95], thus requiring further investigation to specifically analyse the effects of flagellar and spindle defects on chromosomal aberrations in spermatozoa.
Recently, a study has demonstrated a high pregnancy rate and low numerical chromosomal aberration rate as well as good nuclear sperm quality in MMAF patients with DNAH1 mutations [74]. Assisted reproductive technologies have also been used to achieve a successful pregnancy in a few PCD cases. Intracytoplasmic sperm injection was performed in MMAF patients with and without DNAH1 mutations and compared with age-matched non-MMAF patients during the same period. Despite the significant increased rate of disomy XY and chromosome 18, no significant difference in the frequencies of either chromosomes 13, 21, XX or YY disomies or diploidy in sperm from patients with DNAH1 mutation were found compared with non-MMAF patients [74]. Thus, findings from this study suggests that deep genetic investigation using high-throughput sequencing to identify the mutations of genes coding for microtubular proteins and their impact on male infertility are to be focused upon in immediate future.
Mutations of several genes required for sperm development or absence of the respective proteins have been reported to be associated with male infertility by disrupting microtubular functions during formation of sperm head and/or the manchette, and by affecting normal sperm motility [549697]. Deficiencies in the proteins associated with formation of the sperm head (Sept12, SUN, and KASH domain proteins etc.) result in disorganized sperm head formation [98]. Centrobin, an essential centriole protein for centrosome duplication, along with other proteins, such as, ODF1/HSP10, Oazt/Oaz-3, SPATA6 etc. are involved in proper assembly and rigid head-tail conjunction of the sperm [99]. Mutation of these genes causes the acephalic spermatozoa syndrome, fragile necks, and dramatically increased susceptibility to decapitation [97]. A number of proteins, as discussed earlier, is involved in the formation of the manchette (Katnb1, Lrguk1, SUN domain proteins, Hook1, Kif3A etc.). Some mutations of these proteins lead to an elongated and knobbed-like manchette with disorganized microtubules [154754100]. Sperm associated antigen-6 (Spag6), Katnb1, Sept12, Lrguk1, testitin, Ift88, and Kif3A are involved in sperm flagellar motility and maintenance of the structural integrity of mature human sperm. Deficiencies of these proteins cause marked motility defects and morphologically abnormal and disorganized of flagellar structures, including loss of the CP of microtubules and disorganization of the outer dense fibers and fibrous sheath (Table 2) [155496100].
Microtubules play a crucial role in spermatogenesis, as well as in maintenance of sperm morphology and motility. Recent research suggests that abnormalities in microtubular dynamics are associated with male infertility. Till date, numerous genes have been reported to be associated with mitotic/meiotic spindle formation, sperm tail formation, and regulation and trafficking of vesicles within the spermatozoa. However, the proper mechanism of action of each gene and the effects of their abnormalities on male infertility are yet to be explored.
It is important to understand the detailed molecular structure of the microtubules along with their organization and functions. The genes associated with the expressions of different proteins vital to the microtubule functioning, are also to be extensively analyzed. Indepth knowledge about the structure and genetic constitution of the microtubules provide a deeper insight to the pathophysiology of several ciliopathic disorders that affect male fertility and to design targeted therapeutic interventions.
ACKNOWLEDGEMENTS
The authors acknowledge the efforts of Amanda Mendelsohn, Terri O'Brian and Anthony Christovich (Center for Medical Art & Photography, Cleveland Clinic) for drawing the illustrations. Research in this study was supported by funds from American Center for Reproductive Medicine, Cleveland, USA.
References
1. Tuttelmann F, Gromoll J, Kliesch S. Genetics of male infertility. Urologe A. 2008; 47:1561–1562. 1564–1567. PMID: 18953522.
2. Beyaz CC, Gunes S, Onem K, Kulac T, Asci R. Partial deletions of Y-chromosome in infertile men with non-obstructive azoospermia and oligoasthenoteratozoospermia in a Turkish population. In Vivo. 2017; 31:365–371. PMID: 28438864.

3. Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010; 16:231–245. PMID: 19934213.

4. Kaverina I, Straube A. Regulation of cell migration by dynamic microtubules. Semin Cell Dev Biol. 2011; 22:968–974. PMID: 22001384.

5. Watanabe T, Noritake J, Kaibuchi K. Regulation of microtubules in cell migration. Trends Cell Biol. 2005; 15:76–83. PMID: 15695094.

6. Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009; 10:9–20. PMID: 19197328.

7. Linck RW, Chemes H, Albertini DF. The axoneme: the propulsive engine of spermatozoa and cilia and associated ciliopathies leading to infertility. J Assist Reprod Genet. 2016; 33:141–156. PMID: 26825807.

8. Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol. 2015; 16:711–726. PMID: 26562752.

9. O'Donnell L, O'Bryan MK. Microtubules and spermatogenesis. Semin Cell Dev Biol. 2014; 30:45–54. PMID: 24440897.
10. Halpain S, Dehmelt L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006; 7:224. PMID: 16938900.

11. Mohri H, Inaba K, Ishijima S, Baba SA. Tubulin-dynein system in flagellar and ciliary movement. Proc Jpn Acad Ser B Phys Biol Sci. 2012; 88:397–415.

12. Fujita H, Yoshino Y, Chiba N. Regulation of the centrosome cycle. Mol Cell Oncol. 2015; 3:e1075643. PMID: 27308597.

14. Linck RW, Chemes H, Albertini DF. The axoneme: the propulsive engine of spermatozoa and cilia and associated ciliopathies leading to infertility. J Assist Reprod Genet. 2016; 33:141–156. PMID: 26825807.

15. Liu Y, DeBoer K, de Kretser DM, O'Donnell L, O'Connor AE, Merriner DJ, et al. LRGUK-1 is required for basal body and manchette function during spermatogenesis and male fertility. PLoS Genet. 2015; 11:e1005090. PMID: 25781171.

16. Sun X, Kovacs T, Hu YJ, Yang WX. The role of actin and myosin during spermatogenesis. Mol Biol Rep. 2011; 38:3993–4001. PMID: 21107714.

17. Kierszenbaum AL. Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Mol Reprod Dev. 2002; 63:1–4. PMID: 12211054.

18. Lehti MS, Sironen A. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction. 2016; 151:R43–R54. PMID: 26792866.

19. Gob E, Schmitt J, Benavente R, Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS One. 2010; 5:e12072. PMID: 20711465.

20. Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008; 314:1892–1905. PMID: 18396275.

21. Pasch E, Link J, Beck C, Scheuerle S, Alsheimer M. The LINC complex component Sun4 plays a crucial role in sperm head formation and fertility. Biol Open. 2015; 4:1792–1802. PMID: 26621829.

22. Calvi A, Wong AS, Wright G, Wong ES, Loo TH, Stewart CL, et al. SUN4 is essential for nuclear remodeling during mammalian spermiogenesis. Dev Biol. 2015; 407:321–330. PMID: 26417726.

23. Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012; 149:1035–1047. PMID: 22632968.

24. Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, et al. A differential cargo-loading model of ciliary length regulation by IFT. Curr Biol. 2013; 23:2463–2471. PMID: 24316207.

25. Craft JM, Harris JA, Hyman S, Kner P, Lechtreck KF. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J Cell Biol. 2015; 208:223–237. PMID: 25583998.

26. Hayasaka S, Terada Y, Suzuki K, Murakawa H, Tachibana I, Sankai T, et al. Intramanchette transport during primate spermiogenesis: expression of dynein, myosin Va, motor recruiter myosin Va, VIIa-Rab27a/b interacting protein, and Rab27b in the manchette during human and monkey spermiogenesis. Asian J Androl. 2008; 10:561–568. PMID: 18478159.

27. Kierszenbaum AL, Rivkin E, Tres LL, Yoder BK, Haycraft CJ, Bornens M, et al. GMAP210 and IFT88 are present in the spermatid golgi apparatus and participate in the development of the acrosome-acroplaxome complex, head-tail coupling apparatus and tail. Dev Dyn. 2011; 240:723–736. PMID: 21337470.

28. Hamilton JA, Cissen M, Brandes M, Smeenk JM, de Bruin JP, Kremer JA, et al. Total motile sperm count: a better indicator for the severity of male factor infertility than the WHO sperm classification system. Hum Reprod. 2015; 30:1110–1121. PMID: 25788568.

29. Zariwala MA, Leigh MW, Ceppa F, Kennedy MP, Noone PG, Carson JL, et al. Mutations of DNAI1 in primary ciliary dyskinesia: evidence of founder effect in a common mutation. Am J Respir Crit Care Med. 2006; 174:858–866. PMID: 16858015.
30. Sha YW, Ding L, Li P. Management of primary ciliary dyskinesia/Kartagener's syndrome in infertile male patients and current progress in defining the underlying genetic mechanism. Asian J Androl. 2014; 16:101–106. PMID: 24369140.

31. Subramanian R, Kapoor TM. Building complexity: insights into self-organized assembly of microtubule-based architectures. Dev Cell. 2012; 23:874–885. PMID: 23153484.

32. Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003; 112:467–480. PMID: 12600311.

33. Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol. 2005; 17:47–54. PMID: 15661518.

34. Peterman EJ, Scholey JM. Mitotic microtubule crosslinkers: insights from mechanistic studies. Curr Biol. 2009; 19:R1089–R1094. PMID: 20064413.

35. Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol. 2011; 12:709–721. PMID: 21993292.
36. Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009; 10:569–574. PMID: 19513082.

37. Bertalan Z, Budrikis Z, La Porta CA, Zapperi S. Role of the number of microtubules in chromosome segregation during cell division. PLoS One. 2015; 10:e0141305. PMID: 26506005.

38. Helmke KJ, Heald R, Wilbur JD. Interplay between spindle architecture and function. Int Rev Cell Mol Biol. 2013; 306:83–125. PMID: 24016524.

39. Navolanic PM, Sperry AO. Identification of isoforms of a mitotic motor in mammalian spermatogenesis. Biol Reprod. 2000; 62:1360–1369. PMID: 10775188.
40. Tang EI, Mruk DD, Cheng CY. Regulation of microtubule (MT)-based cytoskeleton in the seminiferous epithelium during spermatogenesis. Semin Cell Dev Biol. 2016; 59:35–45. PMID: 26791048.

41. Lagos-Cabre R, Moreno RD. Mitotic, but not meiotic, oriented cell divisions in rat spermatogenesis. Reproduction. 2008; 135:471–478. PMID: 18296512.

42. Holstein AF. Ultrastructural observations on the differentiation of spermatids in man. Andrologia. 1976; 8:157–165. PMID: 962174.

43. Kierszenbaum AL, Tres LL. The acrosome-acroplaxomemanchette complex and the shaping of the spermatid head. Arch Histol Cytol. 2004; 67:271–284. PMID: 15700535.

44. Kierszenbaum AL, Rivkin E, Tres LL. Cytoskeletal track selection during cargo transport in spermatids is relevant to male fertility. Spermatogenesis. 2011; 1:221–230. PMID: 22319670.

45. Eddy EM. The spermatozoon. In : Knobil E, Neill J, editors. Knobil and Neill's physiology of reproduction. St Louis: Acedemc Press;2006. p. 3–54.
46. Zini A, Agarwal A. Spermatogenesis. In : Zini A, Agarwal A, editors. Sperm Chromatin. New York: Springer;2012. p. 19–39.
47. Kramer H, Phistry M. Genetic analysis of hook, a gene required for endocytic trafficking in drosophila. Genetics. 1999; 151:675–684. PMID: 9927460.
48. Baron Gaillard CL, Pallesi-Pocachard E, Massey-Harroche D, Richard F, Arsanto JP, Chauvin JP, et al. Hook2 is involved in the morphogenesis of the primary cilium. Mol Biol Cell. 2011; 22:4549–4562. PMID: 21998199.

49. Schwarz T, Prieler B, Schmid JA, Grzmil P, Neesen J. Ccdc181 is a microtubule-binding protein that interacts with Hook1 in haploid male germ cells and localizes to the sperm tail and motile cilia. Eur J Cell Biol. 2017; 96:276–288. PMID: 28283191.

50. Mendoza-Lujambio I, Burfeind P, Dixkens C, Meinhardt A, Hoyer-Fender S, Engel W, et al. The Hook1 gene is nonfunctional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum Mol Genet. 2002; 11:1647–1658. PMID: 12075009.

51. Kim YH, Haidl G, Schaefer M, Egner U, Mandal A, Herr JC. Compartmentalization of a unique ADP/ATP carrier protein SFEC (Sperm Flagellar Energy Carrier, AAC4) with glycolytic enzymes in the fibrous sheath of the human sperm flagellar principal piece. Dev Biol. 2007; 302:463–476. PMID: 17137571.

52. TAlberts B, Bray D, Lewis J, Raff M, Roberts K, Walter P. Sperm. In : Alberts B, Bray D, Lewis J, Raff M, Roberts K, Walter P, editors. Molecular biology of the cell. New York: Garland Science;2002.
53. Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, et al. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998; 93:277–287. PMID: 9568719.

54. O'Donnell L, Rhodes D, Smith SJ, Merriner DJ, Clark BJ, Borg C, et al. An essential role for katanin p80 and microtubule severing in male gamete production. PLoS Genet. 2012; 8:e1002698. PMID: 22654669.
55. Pleuger C, Fietz D, Hartmann K, Weidner W, Kliesch S, O'Bryan MK, et al. Expression of katanin p80 in human spermatogenesis. Fertil Steril. 2016; 106:1683–1690.e1. PMID: 27717557.

56. Venkatesh D, Mruk D, Herter JM, Cullere X, Chojnacka K, Cheng CY, et al. AKAP9, a regulator of microtubule dynamics, contributes to blood-testis barrier function. Am J Pathol. 2016; 186:270–284. PMID: 26687990.

57. Vogl AW, Weis M, Pfeiffer DC. The perinuclear centriolecontaining centrosome is not the major microtubule organizing center in Sertoli cells. Eur J Cell Biol. 1995; 66:165–179. PMID: 7774603.
58. Cheung K, Senese S, Kuang J, Bui N, Ongpipattanakul C, Gholkar A, et al. Proteomic analysis of the mammalian Katanin family of microtubule-severing enzymes defines Katanin p80 subunit B-like 1 (KATNBL1) as a regulator of mammalian katanin microtubule-severing. Mol Cell Proteomics. 2016; 15:1658–1669. PMID: 26929214.

59. Smith LB, Milne L, Nelson N, Eddie S, Brown P, Atanassova N, et al. KATNAL1 regulation of sertoli cell microtubule dynamics is essential for spermiogenesis and male fertility. PLoS Genet. 2012; 8:e1002697. PMID: 22654668.

60. De Gendt K, Denolet E, Willems A, Daniels VW, Clinckemalie L, Denayer S, et al. Expression of Tubb3, a beta-tubulin isotype, is regulated by androgens in mouse and rat Sertoli cells. Biol Reprod. 2011; 85:934–945. PMID: 21734264.
61. Fedick AM, Eckert K, Thompson K, Forman EJ, Devkota B, Treff NR, et al. Lack of association of KATNAL1 gene sequence variants and azoospermia in humans. J Assist Reprod Genet. 2014; 31:1065–1071. PMID: 24913027.

62. Lobo LJ, Zariwala MA, Noone PG. Primary ciliary dyskinesia. QJM. 2014; 107:691–699. PMID: 24652656.

63. Kartagener M, Horlacher A. Situs viscerum inversus und Polyposis nasi in einem Falle familiaerer Bronchiektasien. Beitr Klin Tuberk. 1935; 87:331–333.
64. Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976; 193:317–319. PMID: 1084576.

65. Barbato A, Frischer T, Kuehni CE, Snijders D, Azevedo I, Baktai G, et al. Primary ciliary dyskinesia: a consensus statement on diagnostic and treatment approaches in children. Eur Respir J. 2009; 34:1264–1276. PMID: 19948909.

66. Kuehni CE, Frischer T, Strippoli MP, Maurer E, Bush A, Nielsen KG, et al. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur Respir J. 2010; 36:1248–1258. PMID: 20530032.

67. Afzelius BA, Eliasson R, Johnsen O, Lindholmer C. Lack of dynein arms in immotile human spermatozoa. J Cell Biol. 1975; 66:225–232. PMID: 1141381.

68. Pedersen H, Rebbe H. Absence of arms in the axoneme of immobile human spermatozoa. Biol Reprod. 1975; 12:541–544. PMID: 1220825.

69. Ji ZY, Sha YW, Ding L, Li P. Genetic factors contributing to human primary ciliary dyskinesia and male infertility. Asian J Androl. 2017; 19:515–520. PMID: 27270341.

70. Alberts B, Bray D, Lewis J, Raff M, Roberts K, Walter P. The self-assembly and dynamic structure of cytoskeletal filaments. In : Alberts B, Bray D, Lewis J, Raff M, Roberts K, Walter P, editors. Molecular biology of the cell. New York: Garland Science;2002.
71. Kurkowiak M, Ziętkiewicz E, Witt M. Recent advances in primary ciliary dyskinesia genetics. J Med Genet. 2015; 52:1–9. PMID: 25351953.

72. Stannard W, Rutman A, Wallis C, O'Callaghan C. Central microtubular agenesis causing primary ciliary dyskinesia. Am J Respir Crit Care Med. 2004; 169:634–637. PMID: 14982824.

73. Chodhari R, Mitchison HM, Meeks M. Cilia, primary ciliary dyskinesia and molecular genetics. Paediatr Respir Rev. 2004; 5:69–76. PMID: 15222957.

74. Wambergue C, Zouari R, Fourati Ben, Martinez G, Devillard F, Hennebicq S, et al. Patients with multiple morphological abnormalities of the sperm flag ella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum Reprod. 2016; 31:1164–1172. PMID: 27094479.
75. Buchdahl RM, Reiser J, Ingram D, Rutman A, Cole PJ, Warner JO. Ciliary abnormalities in respiratory disease. Arch Dis Child. 1988; 63:238–243. PMID: 3355203.

76. Wilton LJ, Teichtahl H, Temple-Smith PD, de Kretser DM. Structural heterogeneity of the axonemes of respiratory cilia and sperm flagella in normal men. J Clin Invest. 1985; 75:825–831. PMID: 3156880.

77. Lee L. Mechanisms of mammalian ciliary motility: insights from primary ciliary dyskinesia genetics. Gene. 2011; 473:57–66. PMID: 21111794.

78. Ishiguro T, Takayanagi N, Hijikata N, Yoshii Y, Yoneda K, Miyahara Y, et al. Primary ciliary dyskinesia. A case report and comparison with 4 previous cases. Nihon Kokyuki Gakkai Zasshi. 2009; 47:242–248. PMID: 19348274.
79. Bartoloni L, Blouin JL, Pan Y, Gehrig C, Maiti AK, Scamuffa N, et al. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc Natl Acad Sci U S A. 2002; 99:10282–10286. PMID: 12142464.

80. Omran H, Haffner K, Volkel A, Kuehr J, Ketelsen UP, Ross UH, et al. Homozygosity mapping of a gene locus for primary ciliary dyskinesia on chromosome 5p and identification of the heavy dynein chain DNAH5 as a candidate gene. Am J Respir Cell Mol Biol. 2000; 23:696–702. PMID: 11062149.
81. Pennarun G, Chapelin C, Escudier E, Bridoux AM, Dastot F, Cacheux V, et al. The human dynein intermediate chain 2 gene (DNAI2): cloning, mapping, expression pattern, and evaluation as a candidate for primary ciliary dyskinesia. Hum Genet. 2000; 107:642–649. PMID: 11153919.

82. Mazor M, Alkrinawi S, Chalifa-Caspi V, Manor E, Sheffield VC, Aviram M, et al. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am J Hum Genet. 2011; 88:599–607. PMID: 21496787.

83. Mitchison HM, Schmidts M, Loges NT, Freshour J, Dritsoula A, Hirst RA, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet. 2012; 44:381–389. s1–s2. PMID: 22387996.

84. Castleman VH, Romio L, Chodhari R, Hirst RA, de Castro SC, Parker KA, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet. 2009; 84:197–209. PMID: 19200523.

85. El Khouri E, Thomas L, Jeanson L, Bequignon E, Vallette B, Duquesnoy P, et al. Mutations in DNAJB13, encoding an HSP40 family member, cause primary ciliary dyskinesia and male infertility. Am J Hum Genet. 2016; 99:489–500. PMID: 27486783.
86. Chemes HE, Olmedo SB, Carrere C, Oses R, Carizza C, Leisner M, et al. Ultrastructural pathology of the sperm flagellum: association between flagellar pathology and fertility prognosis in severely asthenozoospermic men. Hum Reprod. 1998; 13:2521–2526. PMID: 9806277.

87. Baccetti B, Capitani S, Collodel G, Di Cairano G, Gambera L, Moretti E, et al. Genetic sperm defects and consanguinity. Hum Reprod. 2001; 16:1365–1371. PMID: 11425814.

88. Baccetti B, Collodel G, Estenoz M, Manca D, Moretti E, Piomboni P. Gene deletions in an infertile man with sperm fibrous sheath dysplasia. Hum Reprod. 2005; 20:2790–2794. PMID: 15980003.

89. Brown PR, Miki K, Harper DB, Eddy EM. A-kinase anchoring protein 4 binding proteins in the fibrous sheath of the sperm flagellum. Biol Reprod. 2003; 68:2241–2248. PMID: 12606363.

90. Turner RM, Johnson LR, Haig-Ladewig L, Gerton GL, Moss SB. An X-linked gene encodes a major human sperm fibrous sheath protein, hAKAP82. Genomic organization, protein kinase A-RII binding, and distribution of the precursor in the sperm tail. J Biol Chem. 1998; 273:32135–32141. PMID: 9822690.
91. Chemes HE, Rawe VY. The making of abnormal spermatozoa: cellular and molecular mechanisms underlying pathological spermiogenesis. Cell Tissue Res. 2010; 341:349–357. PMID: 20596874.

92. Rawe VY, Olmedo SB, Benmusa A, Shiigi SM, Chemes HE, Sutovsky P. Sperm ubiquitination in patients with dysplasia of the fibrous sheath. Hum Reprod. 2002; 17:2119–2127. PMID: 12151447.

93. Rives NM. Chromosome abnormalities in sperm from infertile men with normal somatic karyotypes: asthenozoospermia. Cytogenet Genome Res. 2005; 111:358–362. PMID: 16192716.

94. Coutton C, Escoffier J, Martinez G, Arnoult C, Ray PF. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update. 2015; 21:455–485. PMID: 25888788.

95. Viville S, Mollard R, Bach ML, Falquet C, Gerlinger P, Warter S. Do morphological anomalies reflect chromosomal aneuploidies?: case report. Hum Reprod. 2000; 15:2563–2566. PMID: 11098027.

96. Teves ME, Sears PR, Li W, Zhang Z, Tang W, van Reesema L, et al. Sperm-associated antigen 6 (SPAG6) deficiency and defects in ciliogenesis and cilia function: polarity, density, and beat. PLoS One. 2014; 9:e107271. PMID: 25333478.

97. Tokuhiro K, Isotani A, Yokota S, Yano Y, Oshio S, Hirose M, et al. OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. PLoS Genet. 2009; 5:e1000712. PMID: 19893612.

98. Zhu F, Wang F, Yang X, Zhang J, Wu H, Zhang Z, et al. Biallelic SUN5 mutations cause autosomal-recessive acephalic spermatozoa syndrome. Am J Hum Genet. 2016; 99:1405.

99. Liska F, Gosele C, Rivkin E, Tres L, Cardoso MC, Domaing P, et al. Rat hd mutation reveals an essential role of centrobin in spermatid head shaping and assembly of the head-tail coupling apparatus. Biol Reprod. 2009; 81:1196–1205. PMID: 19710508.
100. Lehti MS, Kotaja N, Sironen A. KIF3A is essential for sperm tail formation and manchette function. Mol Cell Endocrinol. 2013; 377:44–55. PMID: 23831641.

Fig. 1
The structure and dynamics of microtubule: Microtubules are composed of α- and β-tubulin heterodimers that polymerizes using guanosine-5′-triphosphate (GTP) to form a single proto-filament. The tubulins are arranged in a polarized manner whereby the α-tubulins are exposed at the negative (−) and the β-tubulins at the positive (+) end. The (−) end is more stable, whereas the (+) end can polymerize (grow) and depolymerize (shrink) rapidly, thus rendering microtubules highly dynamic structures. GDP: guanosine 5′-diphosphate.
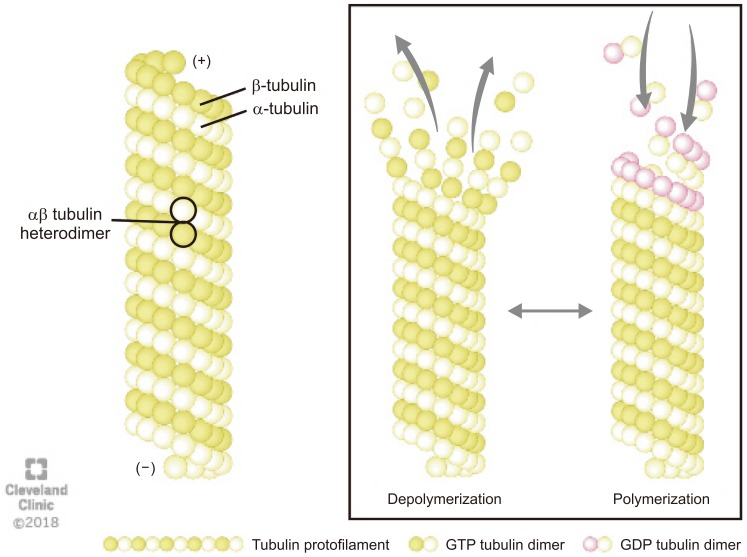
Fig. 2
Schematic diagram of the transport pathways of manchette. Intra-manchette transport (IMT) transfers structural and functional proteins via the microtubule tracks and motor proteins to the basal body region where they are stored. During intra-flagellar transport (IFT), proteins are transported from the sperm cell body to the tip of the flagellum and then back to the sperm head along the axonemal microtubules. Outward or anterograde movement (from the sperm head to the tail) is directly associated with microtubule motor kinesin-2 while inward or retrograde movement is related with dynein 1b. Abnormalities of IMT or IFT microtubule structure could lead to sperm head, neck and tail aberrations because of disruption of the protein delivery to the correct assembly site during spermiogenesis.
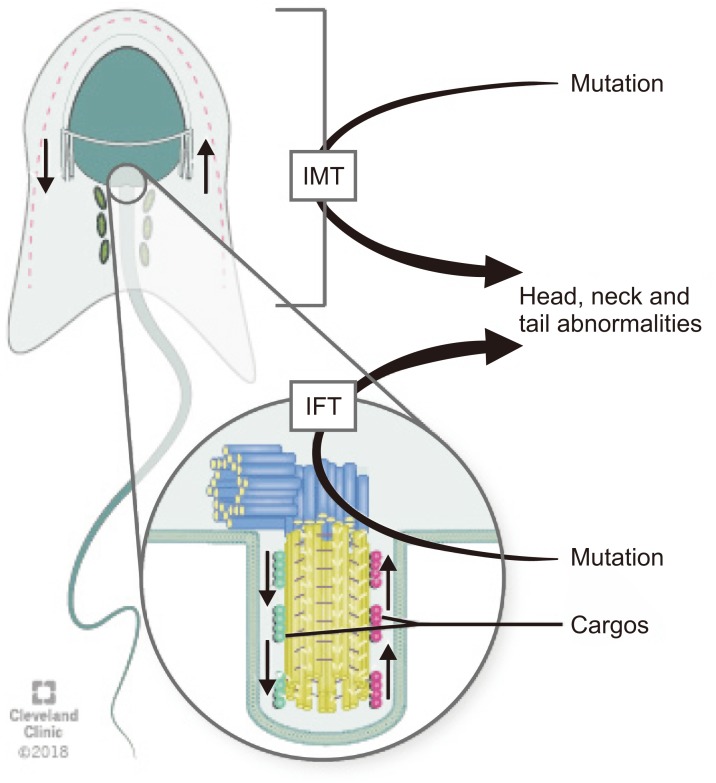
Fig. 3
Schematic representation of flagellum structure. Schematic representation of flagellum structure. A cross-section of flagellum mid-piece shows plasma membrane and mitochondrial sheath surrounding the outer dense fibers. The axoneme displays the characteristic 9+2 arrangement of the microtubules with nine microtubule doublets around the periphery and two singlet microtubules in the center. Adjacent microtubule doublet are connected by Nexin.
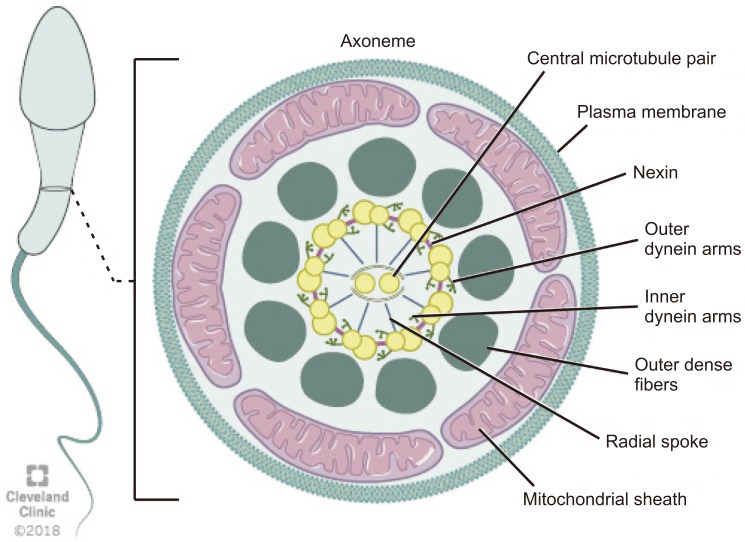
Fig. 5
Abnormalities of the cilia (1) dynein arms abnormalities involve either partial or complete absence of outer dynein arm/inner dynein arm, (2) radial spokes abnormalities including the absence of spokes or deviated central microtubules, (3) inappropriate directionality of cilia, caused by partial or complete absence of the central microtubules, and (4) abnormal number of peripheral microtubules. Atypical arrangements for example 8+1, 8+2, 8+3, or 7+2 have been observed under the microscope on cross-section.
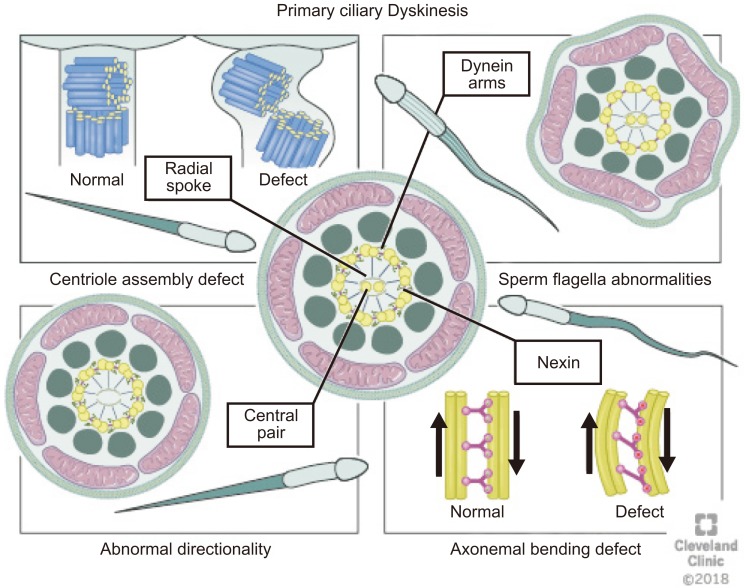
Table 1
Representative mutations of selected genes and their consequences for axonemal structure in human and functional defects in PCD
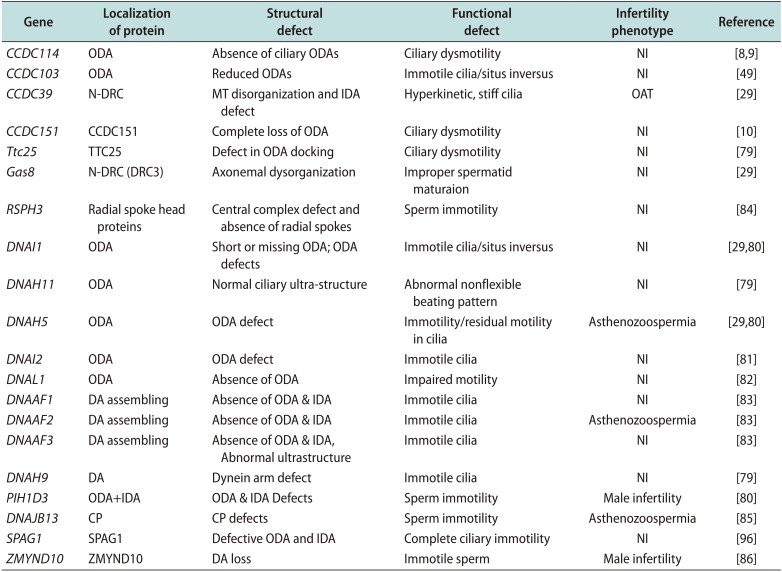
| Gene | Localization of protein | Structural defect | Functional defect | Infertility phenotype | Reference |
|---|---|---|---|---|---|
| CCDC114 | ODA | Absence of ciliary ODAs | Ciliary dysmotility | NI | [89] |
| CCDC103 | ODA | Reduced ODAs | Immotile cilia/situs inversus | NI | [49] |
| CCDC39 | N-DRC | MT disorganization and IDA defect | Hyperkinetic, stiff cilia | OAT | [29] |
| CCDC151 | CCDC151 | Complete loss of ODA | Ciliary dysmotility | NI | [10] |
| Ttc25 | TTC25 | Defect in ODA docking | Ciliary dysmotility | NI | [79] |
| Gas8 | N-DRC (DRC3) | Axonemal dysorganization | Improper maturaion spermatid | NI | [29] |
| RSPH3 | Radial spoke head proteins | Central complex defect and absence of radial spokes | Sperm immotility | NI | [84] |
| DNAI1 | ODA | Short or missing ODA; ODA defects | Immotile cilia/situs inversus | NI | [2980] |
| DNAH11 | ODA | Normal ciliary ultra-structure | Abnormal nonflexible beating pattern | NI | [79] |
| DNAH5 | ODA | ODA defect | Immotility/residual motility in cilia | Asthenozoospermia | [2980] |
| DNAI2 | ODA | ODA defect | Immotile cilia | NI | [81] |
| DNAL1 | ODA | Absence of ODA | Impaired motility | NI | [82] |
| DNAAF1 | DA assembling | Absence of ODA & IDA | Immotile cilia | NI | [83] |
| DNAAF2 | DA assembling | Absence of ODA & IDA | Immotile cilia | Asthenozoospermia | [83] |
| DNAAF3 | DA assembling | Absence of ODA & IDA, Abnormal ultrastructure | Immotile cilia | NI | [83] |
| DNAH9 | DA | Dynein arm defect | Immotile cilia | NI | [79] |
| PIH1D3 | ODA+IDA | ODA & IDA Defects | Sperm immotility | Male infertility | [80] |
| DNAJB13 | CP | CP defects | Sperm immotility | Asthenozoospermia | [85] |
| SPAG1 | SPAG1 | Defective ODA and IDA | Complete ciliary immotility | NI | [96] |
| ZMYND10 | ZMYND10 | DA loss | Immotile sperm | Male infertility | [86] |
Table 2
Genes and proteins associated with manchette abnormalities in mice, and their functions in sperm morphological alterations
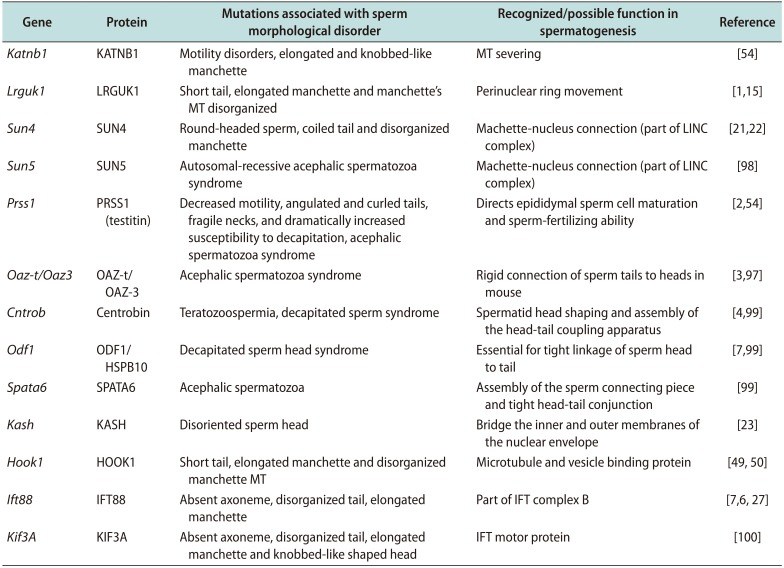
| Gene | Protein | Mutations associated with sperm morphological disorder | Recognized/possible function in spermatogenesis | Reference |
|---|---|---|---|---|
| Katnb1 | KATNB1 | Motility disorders, elongated and knobbed-like manchette | MT severing | [54] |
| Lrguk1 | LRGUK1 | Short tail, elongated manchette and manchette's MT disorganized | Perinuclear ring movement | [115] |
| Sun4 | SUN4 | Round-headed sperm, coiled tail and disorganized manchette | Machette-nucleus connection (part of LINC complex) | [2122] |
| Sun5 | SUN5 | Autosomal-recessive acephalic spermatozoa syndrome | Machette-nucleus connection (part of LINC complex) | [98] |
| Prss1 | PRSS1 (testitin) | Decreased motility, angulated and curled tails, fragile necks, and dramatically increased susceptibility to decapitation, acephalic spermatozoa syndrome | Directs epididymal sperm cell maturation and sperm-fertilizing ability | [254] |
| Oaz-t/Oaz3 | OAZ-t/OAZ-3 | Acephalic spermatozoa syndrome | Rigid connection of sperm tails to heads in mouse | [397] |
| Cntrob | Centrobin | Teratozoospermia, decapitated sperm syndrome | Spermatid head shaping and assembly of | [499] |
| Odf1 | ODF1/HSPB10 | Decapitated sperm head syndrome | Essential for tight linkage of sperm head to tail | [799] |
| Spata6 | SPATA6 | Acephalic spermatozoa | Assembly of the sperm connecting piece and tight head-tail conjunction | [99] |
| Kash | KASH | Disoriented sperm head | Bridge the inner and outer membranes of the nuclear envelope | [23] |
| Hook1 | HOOK1 | Short tail, elongated manchette and disorganized manchette MT | Microtubule and vesicle binding protein | [4950] |
| Ift88 | IFT88 | Absent axoneme, disorganized tail, elongated manchette | Part of IFT complex B | [7627] |
| Kif3A | KIF3A | Absent axoneme, disorganized tail, elongated manchette and knobbed-like shaped head | IFT motor protein | [100] |




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download