Abstract
Humans carry numerous symbiotic microorganisms in their body, most of which are present in the gut. Although recent technological advances have produced extensive research data on gut microbiota, there are various confounding factors (e.g., diet, race, medications) to consider. Sex is one of the important variables affecting the gut microbiota, but the association has not yet been sufficiently investigated. Although the results are inconsistent, several animal and human studies have shown sex differences in gut microbiota. Herein, we review these studies to discuss the sex-dependent differences as well as the possible mechanisms involved.
Numerous microorganisms exist in many sites of the human body, such as the skin, mouth, and vagina, but most of them are in the gut. The term ‘gut microbiota’ refers to all microorganisms present in the gastrointestinal (GI) tract, being composed mostly of bacteria and some viruses and fungi [1]. Approximately 1,000 to 1,150 bacterial species of bacteria are capable of existing in the human colon, and each individual harbors at least 160 species [12]. Because the gut microbiota maintains homeostasis of the host and has functions in immunity and metabolism, the perturbated microbiota (dysbiosis) is associated with not only GI diseases but also other diseases such as cancers, metabolic diseases, allergies, and immunologic disorders [34]. Recent advances in sequencing technology have resulted in a large quantity of data on gut microbiota being produced. It is known that diverse factors, such as diet, race, age, antibiotics, stress, psychological factors, mother's health, mode of delivery, environmental factor, and exercise, can influence the status of the gut microbiota [5]. Therefore, these factors should be considered when planning a study on the microbiota and interpreting the results. Besides these well-known confounding factors, sex or gender might also be an important factor. However, this factor had been ignored by researchers in spite of its importance. Although the results are inconsistent, animal and human studies have shown sex-related differences in gut microbiota [678910111213141516171819]. However, some studies showed no such sex difference [9], and the effect of sex on the gut microbiota appears to be less influential than that of other factors [2021]. In this review, we discuss the animal and human studies on sex differences in the gut microbiota as well as the possible mechanisms involved.
‘Sex’ refers to the biological classification of a species as based on its reproductive systems and the functions derived from a chromosomal type or hormones. The terms ‘male’ and ‘female’ are used when describing the sex of human participants or other sex-related factors [22]. The term ‘gender’ refers to the cultural attitudes and behaviors associated with stereotypical attitudes regarding a person's sex that shape our conceptions of masculinity and femininity [22]. Researchers often confuse these terms and use them indiscriminately even within one article. Because this review deals with the gut microbiota in animal and human studies, we will use the term ‘sex’.
Despite that sex difference can be an important factor not only in human research but also in preclinical research, its importance has only recently been recognized. Most preclinical animal experiments have used male animals, and even some studies did not report the sex of the animal used [23]. Major granting organizations encourage the use of sex or gender as a biological variable regardless of whether an animal study or a clinical trial is being performed [24]. They also recommend reporting the sex or gender of study participants and the sex of animals as well as the origin of cells. The reason for using sex or gender as a variable is due to the issue of reproducibility of the experiment. In this respect, sex analysis should be performed routinely in studies on the gut microbiota, which is still not actively being done despite that such studies are increasing rapidly.
Unexpectedly, the number of human genes coding for proteins found in Human Genomic Projects is only ~20,000, whereas symbiotic bacteria have approximately 100,000 genes, indicating a significant role for them in human physiology [25]. However, we do not know much about gut microbiota yet. Most of the microbial species identified by the molecular method are not cultured or are unculturable [2627]. Similarly, researchers have previously stated that the number of gut microbiota is approximately 1014 cells, which is ten times greater than the number of human body cells. This statement has a long chain of citation originating from a rough estimate of bacteria in fecal contents found in one old literature [28]. A recent study using a new calculation has reported that the number of cells in the gut microbiota are similar to that of human body cells. Interestingly, they showed that the ratio of the numbers of bacterial cells to human cells was different between males and females, being approximately 1.3:1 (38×1012 and 30×1012, respectively) for males and 2.2:1 (44×1012 and 21×1012, respectively) for females [29].
The fetal intestine is supposed to be almost sterile first, then colonized mostly with Lactobacillus species at birth when the fetus passes the mother's vaginal canal, while gut microbiota of babies delivered cesarean section seemed to lack these bacteria. However, the difference in gut microbiota between the birth types seems to be minimized after solid foods are introduced. In addition to the type of birth, the gut microbiota is also influenced by breast-feeding or formula feeding in early life. On the other hand, a recent study reported the presence of bacteria in the placenta or meconium, but it was likely to be the result of contamination [30]. Following the initial colonization of gut bacteria at birth, the species diversity increases and becomes similar to that of an adult when the child is about three years old. The gut microbiota is partly influenced by sex hormones in adolescence and remains stable after that in adulthood. In the elderly of over 70 years of age, the composition of the gut microbiota can be influenced by the changes in intestinal physiological function [3132].
The most potent factor affecting the composition of gut microbiota is the diet. At the species level, the gut microbiota can be divided into three enterotypes according to major microbial community profiles namely, Prevotella, Bacteroides, and Ruminococcus [33]. However, the enterotype is different between Western and Eastern studies [34], and several concerns have been raised to link the discrete enterotypes to human diseases [35].
The factors affecting gut microbiota in the lifelong period are summarized in Fig. 1. As the gut microbiota plays an essential role in the priming and development of the host's immune system, these factors are important in maintaining our health, especially in early life. The normal gut microbiota can be temporarily perturbated by external stressors, such as an infection or antibiotics. Moreover, a fundamental change in living environments, such as immigration or a continuous change in dietary habits, can lead to permanent changes in the gut microbiota [36]. The transient perturbation caused by external stress is usually recovered with time; however, if the degree of such perturbation is too great or takes too long to recover, then dysbiosis will occur [37]. When dysbiosis develops, the normal beneficial function of the gut microbiota is lost, and various disorders could ensue.
Animal studies, mainly in mice, have clearly shown sex-specific differences in the composition of gut microbiota [678383940]. For example, a study with non-obese diabetic (NOD)/ShiLtJ mice demonstrated that the abundance of Porphyromonadaceae, Veillonellaceae, Kineosporiaceae, Peptococcaceae, Enterobacteriaceae, Lactobacillaceae, Cytophagaceae, Peptostreptococcaceae, and Bacteroidaceae at the family level was higher in the male mice than in the female mice [6].
However, some studies failed to find significant sex effects because of the noise introduced by confounding factors, such as diet, age, and host genetic background [8]. It has been suggested that compared to the sex of an animal, the species and strain of animal have more effect on its gut microbiota composition than its sex does [82039].
In a study using two different mouse strains (BALB/c and B6), the males were shown to have a lower microbial diversity and richness than the females, whereas there was no effect of the mouse strain. In contrast, the ratio of Firmicutes/Bacteroidetes was higher in the BALB/c mice, but this ratio was not influenced by sex [39]. In that study, sex and strain explained 11.6% and 26.5% of the variance in microbiota composition, respectively. Sex also influenced the expression profile of genes related to immune functions in the colon in a mouse strain-dependent way [39]. In a study with eight mouse lines from the Collaborative Cross mouse resource, the similarity values were higher between pools of the same line than between pools of the same sex [20].
When the gut microbiota of 89 different inbred mouse strains was analyzed independently [8], clear differences in the gut microbiota composition and diversity were observed between the sexes within each strain, and sex-specific differences were most apparent in the C57BL/6J and C3H/HeJ strains. In a taxonomic analysis, the phyla Actinobacteria and Tenericutes were more abundant in the male mice than in the female mice in the total cohort. At the genus level, Allobaculum, Anaeroplasma, and Erwinia were more abundant in the male mice, whereas SMB53 from family Clostridiaceae and three members of family Lachnospiraceae (Dorea, Coprococcus, and Ruminococcus) were more abundant in the female mice [8]. When the entire population was examined together, both the magnitude and direction of change in the microbiota composition were different between the mouse strains, and no clear differences in patterns between the sexes were observed. These studies suggested that the impact of sex may depend on the host genotype [8].
Although the importance of the gut microbiota to human health has been of interest over several decades, few studies have addressed the effects of sex on the gut microbiota in the human intestines (Table 1) [10111213141516171819]. Two earlier European multinational studies reported conflicting results for the sex differences in gut microbiota composition [910]. In a 2005 study on 91 northern Europeans from France, Denmark, Germany, the Netherlands, and the United Kingdom, which conducted fluorescent in situ hybridization using 18 phylogenetic probes, there were no significant differences in the colonic microbiota between sexes according to principal component analysis [9]. In a 2006 study conducted by four centers in France, Germany, Italy, and Sweden, a higher level of the Bacteroides-Prevotella group was observed in the males [10]. However, two species-specific probes targeting Bacteroides vulgatus and Bacteroides putredinis showed no differences in their abundance between males and females [10]. In a 2008 study of Chinese family members, which was conducted using group-specific DGGE profiling of Bacteroides spp., a higher abundance of Bacteroides thetaiotaomicron was identified in the males [11]. These earlier studies reported minimal information owing to technical issues and a small number of subjects, but recent studies using advanced techniques and large cohorts have provided more detailed results on the sex differences in gut microbiota.
In 2014, researchers who analyzed a 16S rRNA gene sequence data set from the Human Microbiome Project (HMP) Consortium simply reported that sex was associated with the community types identified in the stool, where males were three times more likely to have community type D in which fewer Bacteroides and higher Prevotella were observed [12]. In another large cohort study with two independent, extensively phenotyped cohorts: the Belgian Flemish Gut Flora Project (n=1,106) and the Dutch LifeLines-DEEP study (n=1,135), sex has a 10th effect size among 69 factors that were shown to correlate significantly with overall microbiome community variation [41]. Ever since that HMP analysis report, more studies on sex differences in gut microbiota using 16S rRNA gene sequencing have been reported in recent years. In general, the composition of the gut microbiota seems to be different between sexes, where the α-diversity (i.e., Chao and Shannon) appears to be greater in females. However, the results of each study regarding the differences in microbial taxa between sexes are inconsistent.
In a USA study conducted on a mainly White population, the gut microbiota of the females was characterized by a lower abundance of phylum Bacteroidetes [13]. A study in Italy showed that there were no differences in the lumen-associated microbiota, whereas the mucosa-associated microbiota was different between males and females [16]. The mucosa-associated microbiota in females showed a higher abundance of Actinobacteria, Lactobacillales, Streptococcaceae, and Bifidobacterium and less Veillonellaceae and unclassified Clostridia. At the species level, Gemmiger formicilis was associated with the males and Bifidobacterium adolescentis with the females [16].
In a Spanish study, there were no sex differences in the microbiota diversity, overall composition, phylum level, and the Firmicutes/Bacteroidetes ratio when analyzed without considering the body mass index (BMI) [15]. The finer taxonomic analysis showed a higher abundance of Veillonella and Methanobrevibacter in the males and Bilophila in the females. At the species level, the abundance of Bacteroides caccae was higher in the females, whereas that of Bacteroides plebeius and Coprococcus catus was higher in the males [15]. The sex difference in the microbiota was more obvious when analyzed according to the BMI group in this study [15]. In another study of a large cohort (n=1,135) in the Netherlands, sex was associated with 12 microbial species and 43 metabolic pathways, and the females had a higher abundance of Akkermansia muciniphila even after correcting for all confounding factors, such as diet, lifestyle, and medication [19]. Despite these observations, sex explained only 0.5% of the total variation in the gut microbiota in that study [19].
The studies reported in Asia showed different results from those of the Western studies. In one Japanese study, there was no significant difference in the α-diversity between males and females [18]. That study showed significantly higher levels of Prevotella, Megamonas, Fusobacterium, and Megasphaera in the males, and Bifidobacterium, Ruminococcus, and Akkermansia in the females. A Chinese study reported that there were no overall significant taxonomic differences between males and females. However, at the genus level, Ruminococcus was more abundant in the females [17]. Although a sex difference was observed at the genus level, it was also influenced by the BMI.
There is a report that the sex differences in gut microbiota are more evident when enteric infection is present. In analysis of total gut microbiota from patients with enteric infections (Salmonella, Shiga toxin-producing Escherichia coli, Campylobacter, and Shigella) and of their healthy family members, sex had a significant impact on the overall abundance of taxa, with a slightly higher abundance of Bacteroides in the females and Escherichia in the males [14]. In subgroup analysis, this sex difference in the gut microbiota was not observed in healthy individuals, whereas it was apparent in infected individuals with ten times more different microbial features than that in the healthy individuals [14]. Besides the microbial composition and diversity, a recent study showed that sex was correlated with the functional gene richness of the colon [42].
To discuss the mechanism of sex difference in gut microbiota, it is natural to question whether sex hormones may play a role. As sex differences in gut microbiota do not appear until puberty, the role of sex hormones in shaping the gut microbiota composition is supported [67].
The α-diversity becomes significantly different between males and females after puberty [6]. The microbiota compositions before and after puberty were not different in the female mice, whereas the composition deviated after puberty in the male mice, suggesting male sex hormones may play an important role in the sex differences in gut microbiota in mice. When the androgen source was removed by castration, the gut microbiota of the castrated male was similar to that of a female mouse rather than a male mouse. Furthermore, testosterone treatment after gonadectomy prevented the significant changes in the gut microbiota composition that were observed in the untreated males [8].
Fecal microbiota transplantation (FMT) experiment showed further evidence of the effect of sex difference on shaping the gut microbiota. Even after transplanting the same specific pathogen-free (SPF) feces from a female into male and female germ-free (GF) mice, the gut microbiota after puberty was distinctly segregated according to the sex of the recipient mice [6]. Similarly, a fecal suspension from a 32-year-old woman was administered to male and female GF rats, whereupon the microbiota clustered according to the sex of the host animal despite the same fecal inoculum [43].
There seems to be a reciprocal interaction between gut microbiota and sex hormones. Whereas the level of 17β-estradiol was not different between GF and SPF NOD mice, the level of testosterone was higher in the GF females than in the SPF females and lower in the GF males than in the SPF males [7]. A subset of glycerophospholipid and sphingolipid metabolites was also different between the SPF male and SPF female mice [7]. When the cecal content of a male mouse was administered to weaning GF female mice, the testosterone levels increased in the recipient GF females mice and persisted during the adult stage, but not in GF female mice that received cecal content from a female mouse. This difference disappeared in old age [7]. These results suggest that the commensal bacteria regulate the production and/or utilization of testosterone and cause a difference in metabolism.
Several studies have also suggested the interaction between estrogen and gut microbiota [44]. Bilateral ovariectomy causes microbial dysbiosis in the mouse [845]. In humans, it has been reported that bilateral ovariectomy is related to an increased abundance of Clostridium bolteae [19]. In male and postmenopausal female, but not premenopausal female, the levels of total urinary estrogen were significantly associated with the richness and α-diversity of intestinal microbiota [46]. The higher that the level of non-ovarian systemic estrogens was, the higher was the abundance of fecal Clostridia, including non-Clostridiales and three genera in the Ruminococcaceae family. Interestingly, soy isoflavones, whose metabolites are structurally similar to estrogen, can significantly alter the structure and composition of the intestinal microbial community in the postmenopausal female by increasing the concentration of Bifidobacterium while suppressing unclassified Clostridiaceae [47]. Although the gut microbiota can be modulated by estrogen, gut microbiota can itself also affect the estrogen level by deconjugating the conjugated estrogen molecules excreted through the bile and reabsorbing it through the enterohepatic circulation [46].
In contrast to the gut microbiota in experimental animals growing in a well-controlled environment, the gut microbiota in humans is exposed to various factors, such as foods and medicines in daily life. Because antibiotics, as well as non-antibiotic drugs, have been reported to affect gut microbiota, differences in drug exposure between males and females may be one of the reasons for sex differences in the gut microbiota [48].
In a study of a large cohort in the Netherlands, the males were more likely to be taking drugs for heart disease, whereas the females were more exposed to opiates, laxatives, and antibiotics. Interestingly, the gut microbiota in the females had more antibiotic resistance genes than that in the males [19]. Although it was not consistent with microbial strains showing a sex difference in the entire cohort, drugs that affected the sex hormones altered the gut microbiota. For example, anti-androgen-based oral contraceptives were positively associated with B. caccae and unclassified Coprobacillus strains, and oral contraceptives were associated with an increased abundance of Rothia mucilaginosa [19].
Diet is a most potent modulator of the gut microbiota composition, and animal studies have shown that the interactions between the diet and gut microbiota tended to be sex-dependent, as was observed across different vertebrates, such as wild and laboratory fish, laboratory mice, and humans [849]. The interaction among sex, diet, and gut microbiota also differed between species. It was significant in fish, but not substantial in humans, although there seemed to be a consistent trend that gut microbiota of males was more affected by diet.
The hormonal status affected the microbiota composition more in male mice on a chow diet, whereas this effect was more prevalent in response to a high-fat diet in females [8]. However, the differences between sham control and gonadectomy-treated mice in both sexes were mostly strain-specific. The effect of prebiotics was also different between the sexes. The administration of oligofructose increased the abundance of Bacteroidetes in female rats, but it did not change the microbiota composition in males, even though the butyrate levels were increased [50].
The enterotypes according to the consumed food in humans was reported; however, this study overlooked the differences according to sex [51]. When the existing data were re-analyzed, the correlation of sex and diet affected the abundance of the 125 most common operational taxonomic units, especially those of Fusobacteriaceae [49]. In another study, sex did not have a significant association with the enterotype, but was highly correlated with functional modules, such as enriched aspartate biosynthesis modules in males and enriched inosine monophosphate biosynthesis in females [33].
The underlying mechanism of the sex-dependent interaction between the diet and gut microbiota is still unclear. Fiber intake was related to the gut microbiota composition by reducing the gut transit and pH. In addition, dietary fiber is the main fermentation source used by gut microbiota but has a sex-specific effect [13]. For example, fiber can affect systemic estrogen levels, thereby changing the gut microbiota [52]. Interestingly, there were sex-specific differences in bile acids response to diet. Several bile acids were significantly increased in response to chow diet in gonadectomytreated mice compare to sham-treated mice in both sexes. However, the levels of bile acids in response to a high-fat/high-sucrose diet in gonadectomy-treated male mice is significantly increased compared to that in gonadectomy-treated female mice [8].
Although there exist conflicting results, changes in the body weight were associated with alterations in the proportions of Firmicutes and Bacteroidetes in animal and human studies [5354]. The relationship between the BMI and sex differences in gut microbiota has been reported in Western and Eastern studies. In an USA study, the relationship of the BMI with the overall gut microbiota composition was significant in the females, but not in the males [13]. In the females, lower diversity indices and a lower Bacteroidetes abundance were observed in the overweight and obese subjects relative to the levels in the normal-weight subjects. In a Spanish study, the Firmicutes/Bacteroidetes ratio of the males was higher in the BMI≤33 kg/m2 group and lower in the BMI>33 kg/m2 group compared with that in the females [15]. At the genus level, the abundance of Bacteroides was significantly higher in the females when the BMI was >33 kg/m2 because the abundance of Bacteroides did not change in the females, whereas it decreased in the males with an increase of the BMI [15]. In this study, the microbiota explained 31.17% of the variation in BMI.
In a Chinese study, the α-diversity of gut microbiota was significantly higher in an underweight group than in the other BMI groups, but in the females only. The relative abundance of Fusobacteria was higher in the obese males, whereas that of Actinobacteria was higher in the obese females [17]. At the genus level, the abundance of Bifidobacterium, Coprococcus, and Dialister was higher and that of Phascolarctobacterium was lower in the obese females, whereas the abundance of Fusobacterium was higher in the obese males.
The total body fat content also seemed to influence the diversity and composition of gut microbiota [1755]. Adipose tissue is the main site of extraglandular estrogen synthesis by aromatization of androgens to estrogens, especially in the postmenopausal female [56]. Therefore, it may play an additional role in forming a sex-specific gut microbiota composition.
The colonic transit time is an important factor determining the gut microbiota composition and metabolism. An in vitro study showed that a prolonged transit time decreased the biomass and diversity in the more distal parts of the gut [57]. The stool consistency is strongly correlated with transit time [58]. In patients with constipation, the profile of the fecal microbiota was associated with colonic transit, and genera from Firmicutes (Faecalibacterium, Lactococcus, and Roseburia) correlated with faster colonic transit [59]. The looser stool consistency, which means the faster colonic transit, the lower the richness and the greater the Prevotella enterotype. Conversely, the harder stool consistency, which means the slower colonic transit, the more Ruminococcaceae-Bacteroides enterotype and higher abundance of Methanobrevibacter and Akkermansia populations [58]. There are differences in bowel function and transit between male and female [6061]. In general, females have slower colonic transit time and harder stool form than males [18616263], but there are some reports that it is not different between the sexes [64]. The colonic transit time was shown to be associated with the level of physical activity, where females showed a more significant interaction in this regard [63]. Although not a consistent finding, females sex is a predisposing factor for producing methane gas, which was shown to delay GI motility in an animal model [65]. Taken together, the studies imply that the different colonic transit times between males and females may contribute to sex differences in the gut microbiota.
Sex differences in the development and presentation of various diseases have been known, but the related mechanism is unclear [66]. Sex differences in the gut microbiota may play a role in the sex differences in diseases.
Irritable bowel syndrome (IBS) is a representative disease with sex difference, occurring twice as often in females [67]. In particular, the proportion of females is higher for patients with IBS suffering severe symptoms who visit a tertiary center. The risk of IBS is increased after infectious colitis, and symptoms begin after infectious colitis in about 10% of patients with IBS (so-called post-infectious IBS) [68]. Interestingly, the incidence of post-infectious IBS is higher in females, suggesting that difference in gut microbiota coming between sexes may play an important role in the pathogenesis of post-infectious IBS [68].
The sex differences in the innate and adaptive immune systems are well known [69]. Receptors for sex hormones are expressed on most immune cells, and thus sex hormones may play a role in establishing the sex difference in the immune response [70]. Because the gut microbiota interacts with the host immune system, it can be assumed that the sex differences in gut microbiota have some role in the sex differences in immunity [40]. NOD mice display spontaneous, immune-mediated destruction of their pancreatic beta cells, resulting in type 1 diabetes mellitus. A higher incidence of diabetes mellitus has been reported in female SPF NOD mice than that of males. Interestingly, this sex difference was not observed in GF NOD mice, whereas it appeared again after the colonization of gut microbiota that are known to be associated with the sex difference in SPF NOD mice [6]. In a study of patients with encephalomyelitis/chronic fatigue syndrome, another example of an immune-related disorder, there was no difference in the overall microbiota composition between the sexes. However, the relative abundance Clostridium, Lactobacillus, Streptococcus, and Bifidobacterium according to specific symptom are different between sexes [71]. Intestinal inflammation might also have sex differences in relation to the gut microbiota. In a mouse model of colitis induced with 2,4,6-trinitrobenzenesulfonic acid, the males exhibited more severe colonic inflammation [72]. The FMT animal model of another study showed that female recipients lost significantly more weight after receiving the male microbiota compared with the weight after receiving the female microbiota, suggesting that the male microbiota caused more gut inflammation [40].
Probiotics also elicited different inflammatory responses from female and male mice [73]. In female Wistar rats exposed to water avoidance stress, the administration of Lactobacillus farciminis significantly lowered the colonic mucosal mast cell count and decreased the levels of inflammatory cytokines only in the female rats [73]. In addition, sex differences in response to probiotic Lactobacillus animalis NP-51 administration were reported for cytokine responses, intestinal metabolic profiles and gut microbiota in Mycobacterium-treated mice [74].
The magnitude of the contribution of sex to the gut microbiota is not so clear when compared with other factors such as diet and medication. Nevertheless, the effect of sex difference on the gut microbiota and its interactions with other factors should be routinely analyzed. Moreover, the result should be stated even if there is no main effect of sex in the study. In the past, some drug side effects that occurred preferentially in females were not discovered until after use of the drug in clinical practice, because clinical studies had been mainly performed using male subjects [24]. Similarly, if the sex is not taken into consideration in studies on the gut microbiota, the sex-dependent effect may be overlooked when a microbiota-based therapeutic strategy is available in clinical practice.
There are several points to consider about the studies that have investigated sex differences in the gut microbiota. Sex differences in the gut microbiota composition have been observed mainly in animal studies. It should be also noted that mice studies have shown that the genetic background is a stronger determinant in shaping intestinal microbiota compared to sex difference [20]. Unlike the case with experimental animals, it is difficult to obtain meaningful results on gut microbiota in a human study, because the sex difference might not be easily noticeable and there are many confounding factors, as we have described above [1070].
Another issue is the resolution of currently available methods to investigate gut microbiota. The sex differences in gut microbiota have been observed mainly at the lower taxonomic level; thus current short-read next-generation sequencing (NGS)-based method may have limitations in capturing sex-dependent bacteria [75]. For example, microbes within the same species may produce different metabolites, thus interact differently with sex hormones. Therefore, the full length of 16S rRNA gene or whole genome shotgun sequencing approach may be required to capture the effects of sex differences on gut microbiota, and reveals novel interactions between gut microbiota and sex hormones.
In the future, studies to find the causal relationship among sex, the microbiota, and disease are crucial. To do so, investigations at a finer taxonomic level coupled with multi-omic techniques such as transcriptomics, proteomics, and metabolomics are needed. Not to mention that such studies should be designed to exclude confounding factors as much as possible. These would lead us to treatment strategies that are more tailored to the specific sex for various diseases associated with the gut microbiota.
ACKNOWLEDGEMENTS
The authors thank Professor Moon Young Lee, Department of Physiology, School of Medicine, Wonkwang University for inspiring the creation of this review.
This research was supported by Support Program for Women in Science, Engineering and Technology through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (No. 2019H1C3A1032224).
This research was supported, in part, by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1A6A1A03012862).
References
1. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010; 464:59–65. PMID: 20203603.

2. Sommer F, Bäckhed F. The gut microbiota: masters of host development and physiology. Nat Rev Microbiol. 2013; 11:227–238. PMID: 23435359.
3. Shanahan F. The colonic microbiota in health and disease. Curr Opin Gastroenterol. 2013; 29:49–54. PMID: 23041677.

4. Fukui H, Xu X, Miwa H. Role of gut microbiota-gut hormone axis in the pathophysiology of functional gastrointestinal disorders. J Neurogastroenterol Motil. 2018; 24:367–386. PMID: 29969855.

5. Osadchiy V, Martin CR, Mayer EA. The gut-brain axis and the microbiome: mechanisms and clinical implications. Clin Gastroenterol Hepatol. 2019; 17:322–332. PMID: 30292888.

6. Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013; 39:400–412. PMID: 23973225.

7. Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013; 339:1084–1088. PMID: 23328391.

8. Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016; 7:313–322. PMID: 27355107.

9. Lay C, Rigottier-Gois L, Holmstrøm K, Rajilic M, Vaughan EE, de Vos WM, et al. Colonic microbiota signatures across five northern European countries. Appl Environ Microbiol. 2005; 71:4153–4155. PMID: 16000838.

10. Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006; 72:1027–1033. PMID: 16461645.

11. Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008; 105:2117–2122. PMID: 18252821.

12. Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014; 509:357–360. PMID: 24739969.

13. Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One. 2015; 10:e0124599. PMID: 25874569.

14. Singh P, Manning SD. Impact of age and sex on the composition and abundance of the intestinal microbiota in individuals with and without enteric infections. Ann Epidemiol. 2016; 26:380–385. PMID: 27118420.

15. Haro C, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016; 11:e0154090. PMID: 27228093.

16. Borgo F, Garbossa S, Riva A, Severgnini M, Luigiano C, Benetti A, et al. Body mass index and sex affect diverse microbial niches within the gut. Front Microbiol. 2018; 9:213. PMID: 29491857.

17. Gao X, Zhang M, Xue J, Huang J, Zhuang R, Zhou X, et al. Body mass index differences in the gut microbiota are gender specific. Front Microbiol. 2018; 9:1250. PMID: 29988340.

18. Takagi T, Naito Y, Inoue R, Kashiwagi S, Uchiyama K, Mizushima K, et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J Gastroenterol. 2019; 54:53–63. PMID: 29926167.

19. Sinha T, Vich Vila A, Garmaeva S, Jankipersadsing SA, Imhann F, Collij V, et al. Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes. 2019; 10:358–366. PMID: 30373468.

20. Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb Ecol. 2011; 61:423–428. PMID: 21181142.

21. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486:207–214. PMID: 22699609.
23. Jahng J, Kim YS. Why should we contemplate on gender difference in functional gastrointestinal disorders? J Neurogastroenterol Motil. 2017; 23:1–2. PMID: 28049860.

24. Lee H, Pak YK, Yeo EJ, Kim YS, Paik HY, Lee SK. It is time to integrate sex as a variable in preclinical and clinical studies. Exp Mol Med. 2018; 50:82. PMID: 30038313.

25. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007; 449:804–810. PMID: 17943116.

26. Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012; 18:1185–1193. PMID: 23033984.

27. Bilen M, Dufour JC, Lagier JC, Cadoret F, Daoud Z, Dubourg G, et al. The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome. 2018; 6:94. PMID: 29793532.

28. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans? Cell. 2016; 164:337–340. PMID: 26824647.

29. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016; 14:e1002533. PMID: 27541692.

30. The Lancet Infectious Diseases. Microbiome studies and “blue whales in the Himalayas”. Lancet Infect Dis. 2018; 18:925. PMID: 30152348.
31. Bischoff SC. Microbiota and aging. Curr Opin Clin Nutr Metab Care. 2016; 19:26–30. PMID: 26560527.
32. Salles N. Basic mechanisms of the aging gastrointestinal tract. Dig Dis. 2007; 25:112–117. PMID: 17468545.

33. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011; 473:174–180. PMID: 21508958.

34. Nishijima S, Suda W, Oshima K, Kim SW, Hirose Y, Morita H, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016; 23:125–133. PMID: 26951067.

35. Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, et al. Rethinking “enterotypes”. Cell Host Microbe. 2014; 16:433–437. PMID: 25299329.

36. Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, et al. US immigration westernizes the human gut microbiome. Cell. 2018; 175:962–972.e10. PMID: 30388453.

37. Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017; 15:630–638. PMID: 28626231.

38. Fushuku S, Fukuda K. Gender difference in the composition of fecal flora in laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE). Exp Anim. 2008; 57:489–493. PMID: 18946187.

39. Elderman M, Hugenholtz F, Belzer C, Boekschoten M, van Beek A, de Haan B, et al. Sex and strain dependent differences in mucosal immunology and microbiota composition in mice. Biol Sex Differ. 2018; 9:26. PMID: 29914546.

40. Fransen F, van Beek AA, Borghuis T, Meijer B, Hugenholtz F, van der Gaast-de Jongh C, et al. The impact of gut microbiota on gender-specific differences in immunity. Front Immunol. 2017; 8:754. PMID: 28713378.

41. Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. 2016; 352:560–564. PMID: 27126039.

42. Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016; 352:565–569. PMID: 27126040.

43. Bernbom N, Nørrung B, Saadbye P, Mølbak L, Vogensen FK, Licht TR. Comparison of methods and animal models commonly used for investigation of fecal microbiota: effects of time, host and gender. J Microbiol Methods. 2006; 66:87–95. PMID: 16289391.

44. Chen KL, Madak-Erdogan Z. Estrogen and microbiota cross-talk: Should we pay attention? Trends Endocrinol Metab. 2016; 27:752–755. PMID: 27553057.

45. Cox-York KA, Sheflin AM, Foster MT, Gentile CL, Kahl A, Koch LG, et al. Ovariectomy results in differential shifts in gut microbiota in low versus high aerobic capacity rats. Physiol Rep. 2015; 3:e12488. PMID: 26265751.

46. Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012; 10:253. PMID: 23259758.

47. Nakatsu CH, Armstrong A, Clavijo AP, Martin BR, Barnes S, Weaver CM. Fecal bacterial community changes associated with isoflavone metabolites in postmenopausal women after soy bar consumption. PLoS One. 2014; 9:e108924. PMID: 25271941.

48. Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018; 555:623–628. PMID: 29555994.

49. Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014; 5:4500. PMID: 25072318.

50. Shastri P, McCarville J, Kalmokoff M, Brooks SP, Green-Johnson JM. Sex differences in gut fermentation and immune parameters in rats fed an oligofructose-supplemented diet. Biol Sex Differ. 2015; 6:13. PMID: 26251695.

51. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011; 334:105–108. PMID: 21885731.

52. Gaskins AJ, Mumford SL, Zhang C, Wactawski-Wende J, Hovey KM, Whitcomb BW, et al. Effect of daily fiber intake on reproductive function: the BioCycle study. Am J Clin Nutr. 2009; 90:1061–1069. PMID: 19692496.
53. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005; 102:11070–11075. PMID: 16033867.

54. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006; 444:1022–1023. PMID: 17183309.
55. Mestdagh R, Dumas ME, Rezzi S, Kochhar S, Holmes E, Claus SP, et al. Gut microbiota modulate the metabolism of brown adipose tissue in mice. J Proteome Res. 2012; 11:620–630. PMID: 22053906.

56. Szymczak J, Milewicz A, Thijssen JH, Blankenstein MA, Daroszewski J. Concentration of sex steroids in adipose tissue after menopause. Steroids. 1998; 63:319–321. PMID: 9618794.

57. Tottey W, Feria-Gervasio D, Gaci N, Laillet B, Pujos E, Martin JF, et al. Colonic transit time is a driven force of the gut microbiota composition and metabolism: in vitro evidence. J Neurogastroenterol Motil. 2017; 23:124–134. PMID: 27530163.

58. Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016; 65:57–62. PMID: 26069274.

59. Parthasarathy G, Chen J, Chen X, Chia N, O'Connor HM, Wolf PG, et al. Relationship between microbiota of the colonic mucosa vs feces and symptoms, colonic transit, and methane production in female patients with chronic constipation. Gastroenterology. 2016; 150:367–379.e1. PMID: 26460205.

60. Lampe JW, Fredstrom SB, Slavin JL, Potter JD. Sex differences in colonic function: a randomised trial. Gut. 1993; 34:531–536. PMID: 8387940.

61. Degen LP, Phillips SF. Variability of gastrointestinal transit in healthy women and men. Gut. 1996; 39:299–305. PMID: 8977347.

62. Meier R, Beglinger C, Dederding JP, Meyer-Wyss B, Fumagalli M, Rowedder A, et al. Influence of age, gender, hormonal status and smoking habits on colonic transit time. Neurogastroenterol Motil. 1995; 7:235–238. PMID: 8574912.

63. Song BK, Cho KO, Jo Y, Oh JW, Kim YS. Colon transit time according to physical activity level in adults. J Neurogastroenterol Motil. 2012; 18:64–69. PMID: 22323989.

64. Jung HK, Kim DY, Moon IH. Effects of gender and menstrual cycle on colonic transit time in healthy subjects. Korean J Intern Med. 2003; 18:181–186. PMID: 14619388.

65. Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil. 2014; 20:31–40. PMID: 24466443.

67. Kim YS, Kim N. Sex-gender differences in irritable bowel syndrome. J Neurogastroenterol Motil. 2018; 24:544–558. PMID: 30347934.

68. Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology. 2009; 136:1979–1988. PMID: 19457422.

69. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016; 16:626–638. PMID: 27546235.

70. Elderman M, de Vos P, Faas M. Role of microbiota in sexually dimorphic immunity. Front Immunol. 2018; 9:1018. PMID: 29910797.

71. Wallis A, Butt H, Ball M, Lewis DP, Bruck D. Support for the microgenderome: associations in a human clinical population. Sci Rep. 2016; 6:19171. PMID: 26757840.

72. Kozik AJ, Nakatsu CH, Chun H, Jones-Hall YL. Age, sex, and TNF associated differences in the gut microbiota of mice and their impact on acute TNBS colitis. Exp Mol Pathol. 2017; 103:311–319. PMID: 29175304.

73. Lee JY, Kim N, Nam RH, Sohn SH, Lee SM, Choi D, et al. Probiotics reduce repeated water avoidance stress-induced colonic microinflammation in Wistar rats in a sex-specific manner. PLoS One. 2017; 12:e0188992. PMID: 29244820.

74. Karunasena E, McMahon KW, Chang D, Brashears MM. Host responses to the pathogen Mycobacterium avium subsp. paratuberculosis and beneficial microbes exhibit host sex specificity. Appl Environ Microbiol. 2014; 80:4481–4490. PMID: 24814797.

75. uBiome. A surprising comparison of male vs. female microbiomes [Internet]. San Francisco (CA): uBiome;c2015. cited 2018 Dec 10. Available from: https://ubiome.com/blog/post/surprising-comparison-male-vs-female-microbiomes/.
Fig. 1
Changes in the gut microbiota with time and sex, and the modulatory factors involved. BMI: body mass index.
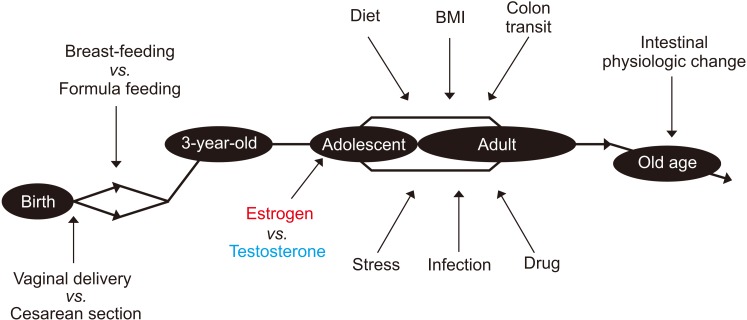
Table 1
Summary of human studies regarding sex differences in gut microbiota
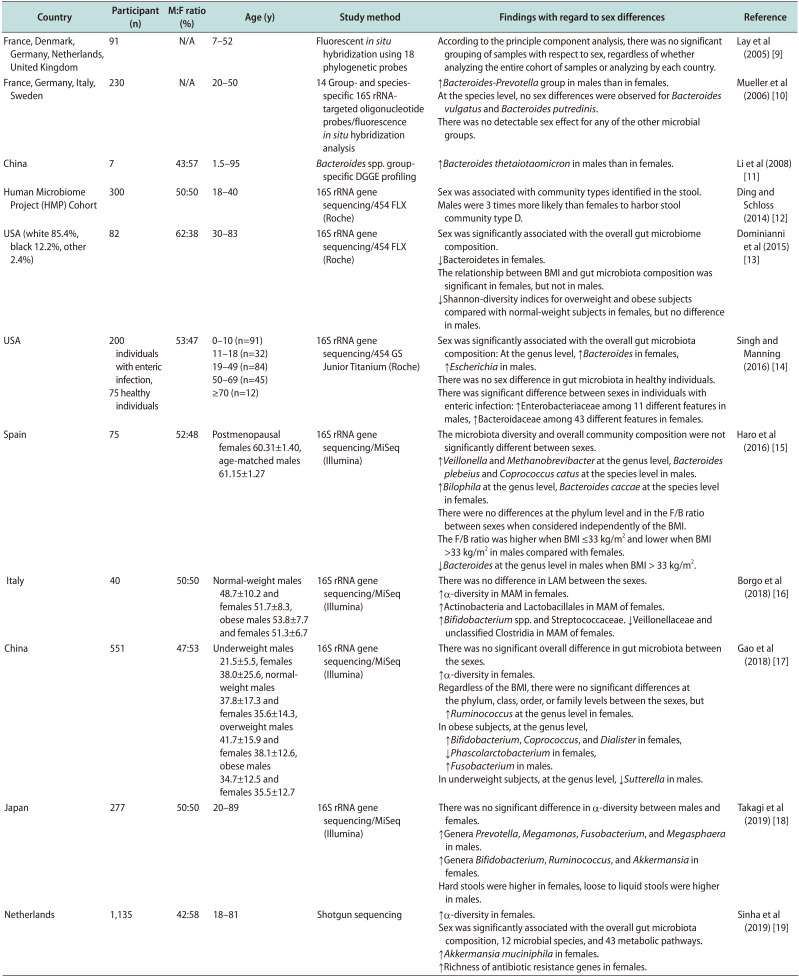
| Country | Participant (n) | M:F ratio (%) | Age (y) | Study method | Findings with regard to sex differences | Reference |
|---|---|---|---|---|---|---|
| France, Denmark, Germany, Netherlands, United Kingdom | 91 | N/A | 7–52 | Fluorescent in situ hybridization using 18 phylogenetic probes | According to the principle component analysis, there was no significant grouping of samples with respect to sex, regardless of whether analyzing the entire cohort of samples or analyzing by each country. | Lay et al (2005) [9] |
| France, Germany, Italy, Sweden | 230 | N/A | 20–50 | 14 Group- and species-specific 16S rRNA-targeted oligonucleotide probes/fluorescence in situ hybridization analysis | ↑Bacteroides-Prevotella group in males than in females. | Mueller et al (2006) [10] |
| At the species level, no sex differences were observed for Bacteroides vulgatus and Bacteroides putredinis. | ||||||
| There was no detectable sex effect for any of the other microbial groups. | ||||||
| China | 7 | 43:57 | 1.5–95 | Bacteroides spp. group-specific DGGE profiling | ↑Bacteroides thetaiotaomicron in males than in females. | Li et al (2008) [11] |
| Human Microbiome Project (HMP) Cohort | 300 | 50:50 | 18–40 | 16S rRNA gene sequencing/454 FLX (Roche) | Sex was associated with community types identified in the stool. | Ding and Schloss (2014) [12] |
| Males were 3 times more likely than females to harbor stool community type D. | ||||||
| USA (white 85.4%, black 12.2%, other 2.4%) | 82 | 62:38 | 30–8 | 16S rRNA gene sequencing/454 FLX (Roche) | Sex was significantly associated with the overall gut microbiome composition. | Dominianni et al (2015) [13] |
| ↓Bacteroidetes in females. | ||||||
| The relationship between BMI and gut microbiota composition was significant in females, but not in males. | ||||||
| ↓Shannon-diversity indices for overweight and obese subjects compared with normal-weight subjects in females, but no difference in males. | ||||||
| USA | 200 individuals with enteric infection, 75 healthy individuals | 53:47 | 0–10 (n=91) | 16S rRNA gene sequencing/454 GS Junior Titanium (Roche) | Sex was significantly associated with the overall gut microbiota composition: At the genus level, ↑Bacteroides in females, ↑Escherichia in males. | Singh and Manning (2016) [14] |
| 11–18 (n=32) | There was no sex difference in gut microbiota in healthy individuals. | |||||
| 19–49 (n=84) | There was significant difference between sexes in individuals with enteric infection: ↑Enterobacteriaceae among 11 different features in males, ↑Bacteroidaceae among 43 different features in females. | |||||
| 50–69 (n=45) | ||||||
| ≥70 (n=12) | ||||||
| Spain | 75 | 52:48 | Postmenopausal females 60.31±1.40, age-matched males 61.15±1.27 | 16S rRNA gene sequencing/MiSeq (Illumina) | The microbiota diversity and overall community composition were not significantly different between sexes. | Haro et al (2016) [15] |
| ↑Veillonella and Methanobrevibacter at the genus level, Bacteroides plebeius and Coprococcus catus at the species level in males. | ||||||
| ↑Bilophila at the genus level, Bacteroides caccae at the species level in females. | ||||||
| There were no differences at the phylum level and in the F/B ratio between sexes when considered independently of the BMI. | ||||||
| The F/B ratio was higher when BMI ≤33 kg/m2 and lower when BMI >33 kg/m2 in males compared with females. | ||||||
| ↓Bacteroides at the genus level in males when BMI > 33 kg/m2. | ||||||
| Italy | 40 | 50:50 | Normal-weight males 48.7±10.2 and females 51.7±8.3, obese males 53.8±7.7 and females 51.3±6.7 | 16S rRNA gene sequencing/MiSeq (Illumina) | There was no difference in LAM between the sexes. | Borgo et al (2018) [16] |
| ↑α-diversity in MAM in females. | ||||||
| ↑Actinobacteria and Lactobacillales in MAM of females. | ||||||
| ↑Bifidobacterium spp. and Streptococcaceae. ↓Veillonellaceae and unclassified Clostridia in MAM of females. | ||||||
| China | 551 | 47:53 | Underweight males 21.5±5.5, females 38.0±25.6, normalweight males 37.8±17.3 and females 35.6±14.3, overweight males 41.7±15.9 and females 38.1±12.6, obese males 34.7±12.5 and females 35.5±12.7 | 16S rRNA gene sequencing/MiSeq (Illumina) | There was no significant overall difference in gut microbiota between the sexes. | Gao et al (2018) [17] |
| ↑α-diversity in females. | ||||||
| Regardless of the BMI, there were no significant differences at the phylum, class, order, or family levels between the sexes, but | ||||||
| ↑Ruminococcus at the genus level in females. | ||||||
| In obese subjects, at the genus level, | ||||||
| ↑Bifidobacterium, Coprococcus, and Dialister in females, | ||||||
| ↓Phascolarctobacterium in females, | ||||||
| ↑Fusobacterium in males. | ||||||
| In underweight subjects, at the genus level, ↓Sutterella in males. | ||||||
| Japan | 277 | 50:50 | 20–89 | 16S rRNA gene sequencing/MiSeq (Illumina) | There was no significant difference in α-diversity between males and females. | Takagi et al (2019) [18] |
| ↑Genera Prevotella, Megamonas, Fusobacterium, and Megasphaera in males. | ||||||
| ↑Genera Bifidobacterium, Ruminococcus, and Akkermansia in females. | ||||||
| Hard stools were higher in females, loose to liquid stools were higher in males. | ||||||
| Netherlands | 1,135 | 42:58 | 18–81 | Shotgun sequencing | Sex was significantly associated with the overall gut microbiota composition, 12 microbial species, and 43 metabolic pathways. | Sinha et al (2019) [19] |
| ↑Akkermansia muciniphila in females. | ||||||
| ↑Richness of antibiotic resistance genes in females. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download