INTRODUCTION
METHODS
Preparation of cells
Transfection of miR-214 into ASCs
Design of animal study
Real-time quantitative PCR
Western blot analysis
Enzyme-linked immunosorbent assay (ELISA)
Special staining including immunohistochemistry
Statistical analysis
Ethics statement
RESULTS
Determination of stability of miR-214-transfected ASCs
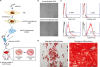 | Fig. 1Determination of stability of miR-214-transfected ASCs. (A) A schematic illustration of study concept. microRNA-214 secretome is obtained from conditioned media in which miR214-transfected ASCs were cultured for 48 hours. Subsequently, we intravenously infused miR-214-secretome into mice with liver fibrosis, and determined the effects of miR-214-secretome on liver fibrosis. (B) Comparison of gross morphology between ASCs either or not transfected with miR-214. Transfecting miR-214 into ASCs did not alter the gross morphology of cultured ASCs. (C) Flow cytometry analysis of expressions of surface markers on ASCs transfected with miR-214. The miR-214-transfected ASCs were negative for CD31 and CD45 (hematopoietic stem cell markers) and positive for CD73 and CD105 (mesenchymal stem cell markers), similar to non-transfected ASCs. (D) Validation of preserved differentiation potential after transfecting ASCs with miR-214. Adipogenic (Left) and osteogenic (Right) differentiation of miR-214-transected ASCs was identified using Oil Red O and Alizarin red stains, respectively (Scale bars = 200 µm). Values are presented as mean ± standard deviation of three independent experiments.ASCs = adipose-derived stem cells, MCM = the secretome released from miR-214-transfected ASCs, HSC = hepatic stellate cell.
*P < 0.05.
|
Antifibrotic effects of the miR-214-secretome in mice with liver fibrosis
 | Fig. 2Antifibrotic effects of the miR-214-secretome in mice with liver fibrosis. The mice with or without liver fibrosis were intravenously infused with normal saline, control secretome, or miR-214-secretome two times a week for 1 week. (A) RT-PCR results demonstrating mRNA expression levels of α-SMA, TGF-β1, and MMP-2 in liver specimens of each group. The infusions of either naïve secretome (CM) or miR-214-secretome (MCM) into mice with liver fibrosis significantly reduced the RNA expression of these markers. miR-214-secretome groups showed significantly reduced expression of MMP-2 compared with naïve secretome group. (B) Results of western blot analysis demonstrating the effects of miR-214-secretome on the expression of various markers in mice with liver fibrosis. The infusion of the miR-214-secretome induced higher expression of PCNA, which reflects hepatocyte proliferation, as well as lower expression of fibrosis-related markers (α-SMA, TIMP-1, TGF-β1, and MMP-2) in the liver specimens than naïve secretome. Values are presented as mean ± standard deviation of three independent experiments.α-SMA = alpha-smooth muscle actin, CM = the secretome obtained from ASCs after 48-hour incubation, MCM = the secretome released from miR-214-transfected ASCs, TGF-β = transforming growth factor-β, MMP-2 = metalloproteinases-2, PCNA = proliferating cell nuclear antigen, TIMP-1 = tissue inhibitor of metalloproteinases-1.
*P < 0.05.
|
Effects of miR-214-secretome on the liver enzymes and systemic inflammation
 | Fig. 3Effects of miR-214-secretome on liver enzymes and systemic inflammation. (A) Serological analysis using blood samples on seventh day post-infusion for the comparison of the serum levels of AST and ALT in each group. Infusion of either kinds of secretome significantly reduced the elevated levels of AST and ALT in the mice with liver fibrosis. Particularly, miR-214-secretome (MCM) significantly decreased serum levels of AST and ALT than naïve secretome (CM). (B) Results of ELISA using blood samples on seventh day post-infusion for the comparison of the serum levels of pro-inflammatory cytokines, such as IL-6 and TNF-α, in each group. In mice with liver fibrosis, secretome infusions significantly reduced serum levels of IL-6 and TNF-α than saline infusion. Of them, miR-214-secretome infusion induced the most significant reduction of these cytokines. Values are presented as mean ± standard deviation of three independent experiments.CM = the secretome obtained from ASCs after 48-hour incubation, MCM = the secretome released from miR-214-transfected ASCs, AST = aspartate transaminase, ALT = alanine transaminase, IL-6 = interleukin 6, TNF-α = tumor necrosis factor-α.
*P < 0.05.
|
Effects of miR-214-secretome on the histology of the liver
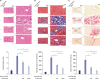 | Fig. 4Effects of miR-214-secretome on the histological characteristics of the liver specimens. (A) Top: H&E stains of the liver specimens in each group. Although inflammation driven by TAA deranged the histologic architectures of the liver, infusion of either kinds of secretomes significantly recovered it. Of the two secretome groups, histological improvement appeared to be more prominent in miR-214-secretome (MCM) group than in naïve secretome (CM) group. Bottom: The severity of liver injury was assessed using Suzuki classification. (B) Top: Masson's trichrome demonstrating the superior antifibrotic activities of secretome infusions. Of the two secretome groups, miR-214-secretome group showed significantly higher amelioration of fibrosis than naïve secretome group. Bottom: Relative densities of these markers quantified using ImageJ software. (C) Top: Sirius red stains demonstrating the superior antifibrotic activities of secretome infusions. Of the two secretome groups, miR-214-secretome group showed significantly higher amelioration of fibrosis than control secretome group. Bottom: Relative densities of these markers quantified using ImageJ software. Values are presented as mean ± standard deviation of three independent experiments.MCM = the secretome released from miR-214-transfected ASCs, CM = the secretome obtained from ASCs after 48-hour incubation, H&E = hematoxylin and eosin, TAA = thioacetamide.
*P < 0.05.
|
Effects of miR-214-secretome on immunohistochemical staining of the liver
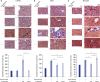 | Fig. 5Effects of miR-214-secretome on immunohistochemical staining of the liver. (A) Top: α-SMA IHC staining showing that infusion of miR-214-secretome (MCM) significantly reduced the expression of α-SMA compared to saline infusion. Bottom: Relative densities of these markers quantified using ImageJ software. (B) Top: SOD IHC staining showing that infusion of miR-214-secretome significantly increased the expression of SOD compared to that by saline infusion. Bottom: Relative densities of these markers quantified using ImageJ software. (C) Top: Albumin IHC staining showing that infusion of miR-214-secretome significantly increased the expression of albumin compared to saline infusion. Bottom: Relative densities of these markers quantified using ImageJ software. Values are presented as mean ± standard deviation of three independent experiments.α-SMA = alpha-smooth muscle actin, MCM = the secretome released from miR-214-transfected ASCs, CM = the secretome obtained from ASCs after 48-hour incubation, SOD = superoxide dismutase, IHC = immunohistochemistry.
*P < 0.05.
|




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download