Abstract
The commensal microbiota contributes to the maintenance of immune homeostasis in the human body. Autoimmunity can be aggravated or alleviated by the microbiota, which affects both innate and adaptive immune cells. Many studies have demonstrated the role of gut dysbiosis, the alteration of the gut microbiome, in the development and progression of numerous autoimmune diseases. Systemic sclerosis (SSc) is an autoimmune disease of the connective tissue and is characterized by skin and lung fibrosis, as well as injuries in small arteries. Recent studies have shown variable degrees of dysbiosis in SSc patients and the effect of probiotics on these patients, providing evidence for the potential link between microbiota and SSc. However, further research is needed to elucidate the key microorganisms and the mechanisms through which they affect the pathoimmuno-logical process of SSc. This review summarizes the current knowledge regarding the association between microbiota and SSc, and discusses the changing perspectives and potential therapy strategies based on the microbiota and its products.
Go to : 
REFERENCES
1. De Luca F, Shoenfeld Y. The microbiome in autoimmune diseases. Clin Exp Immunol. 2019; 195:74–85.

2. Opazo MC, Ortega-Rocha EM, Coronado-Arrázola I, Bonifaz LC, Boudin H, Neunlist M, et al. Intestinal microbiota influences non-intestinal related autoimmune diseases. Front Microbiol. 2018; 9:432.

3. Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015; 159:122–7.

4. Bellocchi C, Volkmann ER. Update on the gastrointestinal microbiome in systemic sclerosis. Curr Rheumatol Rep. 2018; 20:49.

5. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011; 332:974–7.

6. Dasgupta S, Erturk-Hasdemir D, Ochoa-Reparaz J, Reinecker HC, Kasper DL. Plasmacytoid dendritic cells mediate antiinflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 2014; 15:413–23.

7. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013; 500:232–6.

8. Hayashi A, Sato T, Kamada N, Mikami Y, Matsuoka K, Hisamatsu T, et al. A single strain of Clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe. 2013; 13:711–22.

9. Cavaglieri CR, Nishiyama A, Fernandes LC, Curi R, Miles EA, Calder PC. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci. 2003; 73:1683–90.

10. Zhong D, Wu C, Zeng X, Wang Q. The role of gut microbiota in the pathogenesis of rheumatic diseases. Clin Rheumatol. 2018; 37:25–34.

11. Zhang M, Zhou Q, Dorfman RG, Huang X, Fan T, Zhang H, et al. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol. 2016; 16:84.

12. Mizuno M, Noto D, Kaga N, Chiba A, Miyake S. The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS One. 2017; 12:e0173032.

13. Pozuelo M, Panda S, Santiago A, Mendez S, Accarino A, Santos J, et al. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci Rep. 2015; 5:12693.

14. Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009; 15:1183–9.

15. Hevia A, Milani C, López P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio. 2014; 5:e01548–14.

16. Rojo D, Hevia A, Bargiela R, López P, Cuervo A, González S, et al. Ranking the impact of human health disorders on gut metabolism: systemic lupus erythematosus and obesity as study cases. Sci Rep. 2015; 5:8310.

17. Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016; 8:43.

18. Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016; 7:12015.

19. Freedman SN, Shahi SK, Mangalam AK. The “gut feeling”: breaking down the role of gut microbiome in multiple sclerosis. Neurotherapeutics. 2018; 15:109–25.

20. López P, de Paz B, Rodríguez-Carrio J, Hevia A, Sánchez B, Margolles A, et al. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep. 2016; 6:24072.

21. Oliveira Mde F, Rocha Bde O, Duarte GV. Psoriasis: classical and emerging comorbidities. An Bras Dermatol. 2015; 90:9–20.
22. Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015; 67:128–39.

23. Emmanuel A. Current management of the gastrointestinal complications of systemic sclerosis. Nat Rev Gastroenterol Hepatol. 2016; 13:461–72.

24. Patrone V, Puglisi E, Cardinali M, Schnitzler TS, Svegliati S, Festa A, et al. Gut microbiota profile in systemic sclerosis patients with and without clinical evidence of gastrointestinal involvement. Sci Rep. 2017; 7:14874.

25. Volkmann ER, Hoffmann-Vold AM, Chang YL, Jacobs JP, Tillisch K, Mayer EA, et al. Systemic sclerosis is associated with specific alterations in gastrointestinal microbiota in two independent cohorts. BMJ Open Gastroenterol. 2017; 4:e000134.

26. Varga J, Trojanowska M, Kuwana M. Pathogenesis of systemic sclerosis: recent insights of molecular and cellular mechanisms and therapeutic opportunities. J Scleroderma Relat Disord. 2017; 2:137–52.

27. Chaudhuri V, Zhou L, Karasek M. Inflammatory cytokines induce the transformation of human dermal microvascular endothelial cells into myofibroblasts: a potential role in skin fibrogenesis. J Cutan Pathol. 2007; 34:146–53.

28. Chrobak I, Lenna S, Stawski L, Trojanowska M. Interferon-γ promotes vascular remodeling in human microvascular endothelial cells by upregulating endothelin (ET)-1 and transforming growth factor (TGF) β 2. J Cell Physiol. 2013; 228:1774–83.
29. Echeverría C, Montorfano I, Sarmiento D, Becerra A, Nuñez-Villena F, Figueroa XF, et al. Lipopolysaccharide induces a fibrotic-like phenotype in endothelial cells. J Cell Mol Med. 2013; 17:800–14.

30. Mahler GJ, Farrar EJ, Butcher JT. Inflammatory cytokines promote mesenchymal transformation in embryonic and adult valve endothelial cells. Arterioscler Thromb Vasc Biol. 2013; 33:121–30.

31. Maleszewska M, Moonen JR, Huijkman N, van de Sluis B, Krenning G, Harmsen MC. IL-1β and TGFβ 2 synergistically induce endothelial to mesenchymal transition in an NF κB-dependent manner. Immunobiology. 2013; 218:443–54.
32. Piera-Velazquez S, Mendoza FA, Jimenez SA. Endothelial to mesenchymal transition (EndoMT) in the pathogenesis of human fibrotic diseases. J Clin Med. 2016; 5:E45.

33. Stawski L, Han R, Bujor AM, Trojanowska M. Angiotensin II induces skin fibrosis: a novel mouse model of dermal fibrosis. Arthritis Res Ther. 2012; 14:R194.

34. Wermuth PJ, Li Z, Mendoza FA, Jimenez SA. Stimulation of transforming growth factor-β 1-induced endothelial-to-mesenchymal transition and tissue fibrosis by endothelin-1 (ET-1): a novel profibrotic effect of ET-1. PLoS One. 2016; 11:e0161988.
35. Akamatsu T, Arai Y, Kosugi I, Kawasaki H, Meguro S, Sakao M, et al. Direct isolation of myofibroblasts and fibroblasts from bleomycin-injured lungs reveals their functional similarities and differences. Fibrogenesis Tissue Repair. 2013; 6:15.

36. Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, et al. Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science. 2015; 348:aaa2151.

37. Wermuth PJ, Carney KR, Mendoza FA, Piera-Velazquez S, Jimenez SA. Endothelial cell-specific activation of transforming growth factor-β signaling in mice induces cutaneous, visceral, and microvascular fibrosis. Lab Invest. 2017; 97:806–18.

38. Kitaba S, Murota H, Terao M, Azukizawa H, Terabe F, Shima Y, et al. Blockade of interleukin-6 receptor alleviates disease in mouse model of scleroderma. Am J Pathol. 2012; 180:165–76.

39. Fuschiotti P, Medsger TA Jr, Morel PA. Effector CD8+ T cells in systemic sclerosis patients produce abnormally high levels of interleukin-13 associated with increased skin fibrosis. Arthritis Rheum. 2009; 60:1119–28.

41. O'Reilly S. Role of interleukin-13 in fibrosis, particularly systemic sclerosis. Biofactors. 2013; 39:593–6.
42. Lakos G, Melichian D, Wu M, Varga J. Increased bleomy-cin-induced skin fibrosis in mice lacking the Th1-specific transcription factor T-bet. Pathobiology. 2006; 73:224–37.

43. Wohlfahrt T, Rauber S, Uebe S, Luber M, Soare A, Ekici A, et al. PU.1 controls fibroblast polarization and tissue fibrosis. Nature. 2019; 566:344–9.

44. Hua-Huy T, Dinh-Xuan AT. Cellular and molecular mechanisms in the pathophysiology of systemic sclerosis. Pathol Biol (Paris). 2015; 63:61–8.

45. Laurent P, Sisirak V, Lazaro E, Richez C, Duffau P, Blanco P, et al. Innate immunity in systemic sclerosis fibrosis: recent advances. Front Immunol. 2018; 9:1702.

46. Fang F, Marangoni RG, Zhou X, Yang Y, Ye B, Shangguang A, et al. Toll-like receptor 9 signaling is augmented in systemic sclerosis and elicits transforming growth factor β-dependent fibroblast activation. Arthritis Rheumatol. 2016; 68:1989–2002.

47. Fang F, Ooka K, Sun X, Shah R, Bhattacharyya S, Wei J, et al. A synthetic TLR3 ligand mitigates profibrotic fibroblast responses by inducing autocrine IFN signaling. J Immunol. 2013; 191:2956–66.

48. O'Reilly S, Cant R, Ciechomska M, Finnigan J, Oakley F, Hambleton S, et al. Serum amyloid A induces interleukin-6 in dermal fibroblasts via Toll-like receptor 2, interleukin-1 receptor-associated kinase 4 and nuclear factor-κ B. Immunology. 2014; 143:331–40.
49. Stifano G, Christmann RB. Macrophage involvement in systemic sclerosis: do we need more evidence? Curr Rheumatol Rep. 2016; 18:2.

50. Frantz C, Pezet S, Avouac J, Allanore Y. Soluble CD163 as a potential biomarker in systemic sclerosis. Dis Markers. 2018; 2018; 8509583.

51. Taroni JN, Greene CS, Martyanov V, Wood TA, Christmann RB, Farber HW, et al. A novel multi-network approach reveals tissue-specific cellular modulators of fibrosis in systemic sclerosis. Genome Med. 2017; 9:27.

52. Knipper JA, Willenborg S, Brinckmann J, Bloch W, Maaß T, Wagener R, et al. Interleukin-4 receptor α signaling in myeloid cells controls collagen fibril assembly in skin repair. Immunity. 2015; 43:803–16.

53. Canesso MC, Vieira AT, Castro TB, Schirmer BG, Cisalpino D, Martins FS, et al. Skin wound healing is accelerated and scarless in the absence of commensal microbiota. J Immunol. 2014; 193:5171–80.

54. Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Núñez G, Shibuya A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe. 2014; 15:95–102.

55. Ah Kioon MD, Tripodo C, Fernandez D, Kirou KA, Spiera RF, Crow MK, et al. Plasmacytoid dendritic cells promote systemic sclerosis with a key role for TLR8. Sci Transl Med. 2018; 10:eaam8458.

56. Truchetet ME, Brembilla NC, Montanari E, Allanore Y, Chizzolini C. Increased frequency of circulating Th22 in addition to Th17 and Th2 lymphocytes in systemic sclerosis: association with interstitial lung disease. Arthritis Res Ther. 2011; 13:R166.

57. Dumoitier N, Chaigne B, Régent A, Lofek S, Mhibik M, Dorfmüller P, et al. Scleroderma peripheral B lymphocytes secrete interleukin-6 and transforming growth factor β and activate fibroblasts. Arthritis Rheumatol. 2017; 69:1078–89.

58. Hinchcliff M, Huang CC, Wood TA, Matthew Mahoney J, Martyanov V, Bhattacharyya S, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol. 2013; 133:1979–89.

59. Johnson ME, Franks JM, Cai G, Mehta BK, Wood TA, Archambault K, et al. Microbiome dysbiosis is associated with disease duration and increased inflammatory gene expression in systemic sclerosis skin. Arthritis Res Ther. 2019; 21:49.

60. Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One. 2008; 3:e2696.

61. Pendergrass SA, Lemaire R, Francis IP, Mahoney JM, Lafyatis R, Whitfield ML. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012; 132:1363–73.

62. Volkmann ER, Chang YL, Barroso N, Furst DE, Clements PJ, Gorn AH, et al. Association of systemic sclerosis with a unique colonic microbial consortium. Arthritis Rheumatol. 2016; 68:1483–92.

63. Lim LH, Li HY, Huang CH, Lee BW, Lee YK, Chua KY. The effects of heat-killed wild-type Lactobacillus casei Shirota on allergic immune responses in an allergy mouse model. Int Arch Allergy Immunol. 2009; 148:297–304.
64. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum Increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κ B, and upregulating expression of microRNA-21. Gastroenterology. 2017; 152:851–66.e24.
65. Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016; 68:2646–61.

66. Kugathasan S, Denson LA, Walters TD, Kim MO, Marigorta UM, Schirmer M, et al. Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet. 2017; 389:1710–8.
67. Bellocchi C, Fernández-Ochoa Á, Montanelli G, Vigone B, Santaniello A, Milani C, et al. Microbial and metabolic mul-ti-omic correlations in systemic sclerosis patients. Ann N Y Acad Sci. 2018; 1421; 97–109.

68. Andréasson K, Alrawi Z, Persson A, Jönsson G, Marsal J. Intestinal dysbiosis is common in systemic sclerosis and associated with gastrointestinal and extraintestinal features of disease. Arthritis Res Ther. 2016; 18:278.

69. Marighela TF, Arismendi MI, Marvulle V, Brunialti MKC, Salomão R, Kayser C. Effect of probiotics on gastrointestinal symptoms and immune parameters in systemic sclerosis: a randomized placebo-controlled trial. Rheumatology (Oxford). 2019 May 5; [Epub].DOI: DOI:10.1093/rheumatology/kez160.

70. Park JS, Choi JW, Jhun J, Kwon JY, Lee BI, Yang CW, et al. Lactobacillus acidophilus improves intestinal inflammation in an acute colitis mouse model by regulation of Th17 and Treg cell balance and fibrosis development. J Med Food. 2018; 21:215–24.

71. Frech TM, Khanna D, Maranian P, Frech EJ, Sawitzke AD, Murtaugh MA. Probiotics for the treatment of systemic sclerosis-associated gastrointestinal bloating/ distention. Clin Exp Rheumatol. 2011; 29(2 Suppl 65):S22–5.
72. Enteshari-Moghaddam A, Movassaghi S, Rostamian A. Effect of probiotics in the treatment of gastrointestinal symptoms in patients with scleroderma. Int J Sci Rep. 2016; 2:94–8.

73. de Oliveira GLV, Leite AZ, Higuchi BS, Gonzaga MI, Mariano VS. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology. 2017; 152:1–12.

74. Owaga E, Hsieh RH, Mugendi B, Masuku S, Shih CK, Chang JS. Th17 cells as potential probiotic therapeutic targets in inflammatory bowel diseases. Int J Mol Sci. 2015; 16:20841–58.
75. Mano MCR, Neri-Numa IA, da Silva JB, Paulino BN, Pessoa MG, Pastore GM. Oligosaccharide biotechnology: an approach of prebiotic revolution on the industry. Appl Microbiol Biotechnol. 2018; 102:17–37.

76. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017; 14:491–502.

77. Volkmann ER. Intestinal microbiome in scleroderma: recent progress. Curr Opin Rheumatol. 2017; 29:553–60.

78. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012; 488:178–84.

79. Guo X, Li J, Tang R, Zhang G, Zeng H, Wood RJ, et al. High fat diet alters gut microbiota and the expression of paneth cell-antimicrobial peptides preceding changes of circulating inflammatory cytokines. Mediators Inflamm. 2017; 2017; 9474896.

80. Hong J, Jia Y, Pan S, Jia L, Li H, Han Z, et al. Butyrate alleviates high fat diet-induced obesity through activation of adiponectin-mediated pathway and stimulation of mitochondrial function in the skeletal muscle of mice. Oncotarget. 2016; 7:56071–82.

81. Staudacher HM, Irving PM, Lomer MC, Whelan K. Mechanisms and efficacy of dietary FODMAP restriction in IBS. Nat Rev Gastroenterol Hepatol. 2014; 11:256–66.

82. McIntosh K, Reed DE, Schneider T, Dang F, Keshteli AH, De Palma G, et al. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut. 2017; 66:1241–51.

83. Marie I, Leroi AM, Gourcerol G, Levesque H, Ménard JF, Ducrotte P. Fructose malabsorption in systemic sclerosis. Medicine (Baltimore). 2015; 94:e1601.

84. Denton CP, Murray C. Cause or effect? Interpreting emerging evidence for dysbiosis in systemic sclerosis. Arthritis Res Ther. 2019; 21:81.

85. Rosman Y, Lidar M, Shoenfeld Y. Antibiotic therapy in autoimmune disorders. Clin Pract. 2014; 11:91–103.

86. Mu Q, Tavella VJ, Kirby JL, Cecere TE, Chung M, Lee J, et al. Antibiotics ameliorate lupus-like symptoms in mice. Sci Rep. 2017; 7:13675.

87. Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016; 8:39.

88. Mehta H, Goulet PO, Mashiko S, Desjardins J, Pérez G, Koenig M, et al. Early-life antibiotic exposure causes intestinal dysbiosis and exacerbates skin and lung pathology in experimental systemic sclerosis. J Invest Dermatol. 2017; 137:2316–25.

90. Weyrich LS, Dixit S, Farrer AG, Cooper AJ, Cooper AJ. The skin microbiome: associations between altered microbial communities and disease. Australas J Dermatol. 2015; 56:268–74.

92. Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009; 15:1377–82.

93. Arron ST, Dimon MT, Li Z, Johnson ME, Wood TA, Feeney L, et al. High Rhodotorula sequences in skin transcriptome of patients with diffuse systemic sclerosis. J Invest Dermatol. 2014; 134:2138–45.

94. Schwarz A, Bruhs A, Schwarz T. The short-chain fatty acid sodium butyrate functions as a regulator of the skin immune system. J Invest Dermatol. 2017; 137:855–64.

Go to : 
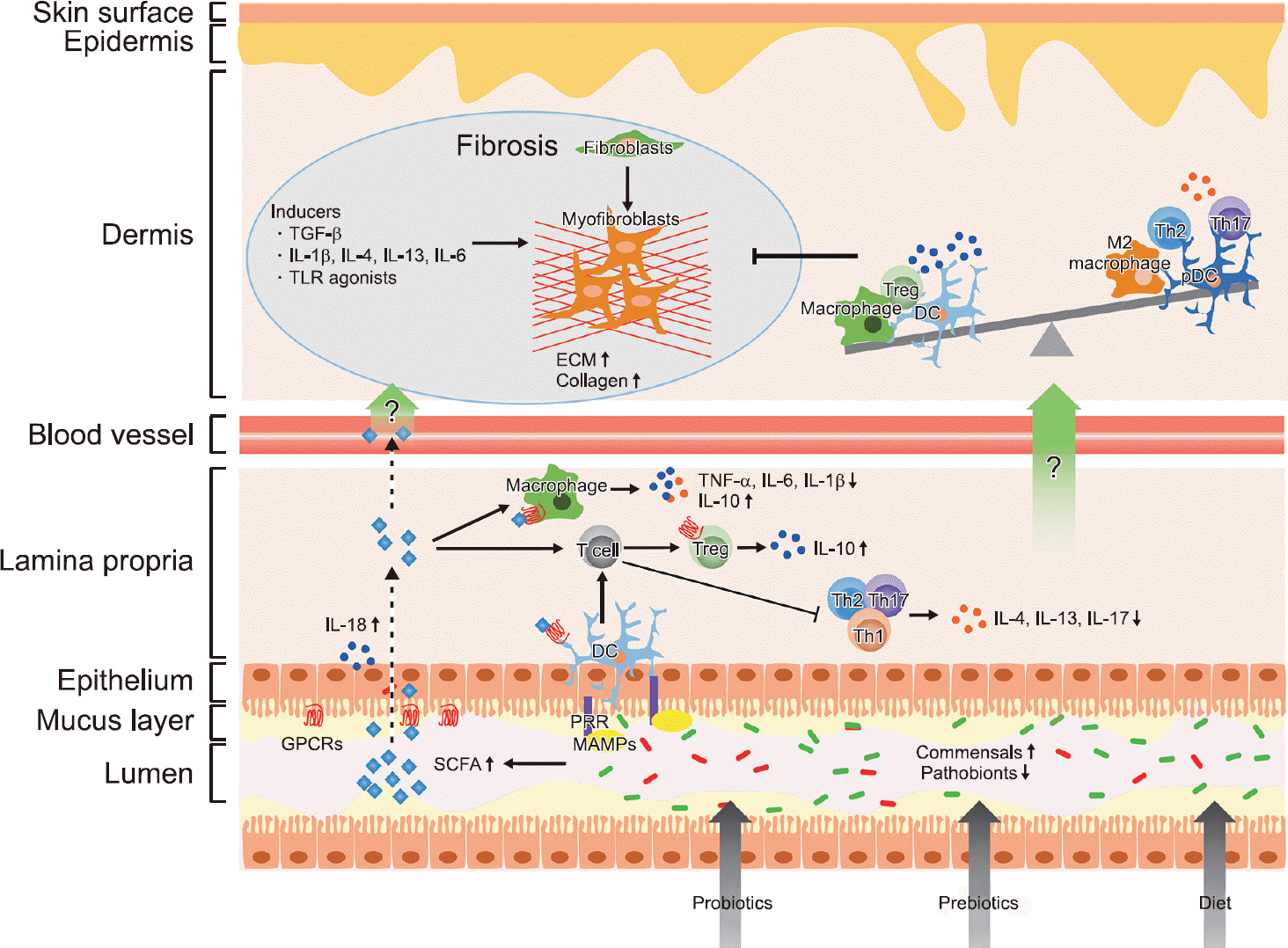 | Figure 1.Potential effects of microbiota-targeted challenges in systemic sclerosis (SSc). Probiotics, prebiotics, and dietary fibers can help produce microbial metabolites, such as short-chain fatty acids (SCFAs), via the recovery of commensal bacteria. SCFAs bind to G protein-coupled receptors (GPCRs) on intestinal epithelial cells and immune cells. In turn, SCFAs regulate intestinal barrier integrity by inducing secretion of interleukin (IL)-18 by epithelial cells, differentiation of T cells into regulatory T (Treg) cells, and proinflammatory cytokine production by macrophages. Dendritic cells (DCs) also regulate T cell differentiation by both SCFAs and microbe-associated molecular patterns (MAMPs) sensing through pattern-recognition receptors (PRRs). Presumably, the circulation of SCFAs and anti-inflammatory cytokines might prevent fibrosis by modulating the dysregulated immune system in SSc. TGF-β: transforming growth factor-β, TLR: toll-like receptor, ECM: extracellular matrix, pDC: plasmacytoid DC, TNF-α: tumor necrosis factor-α. |
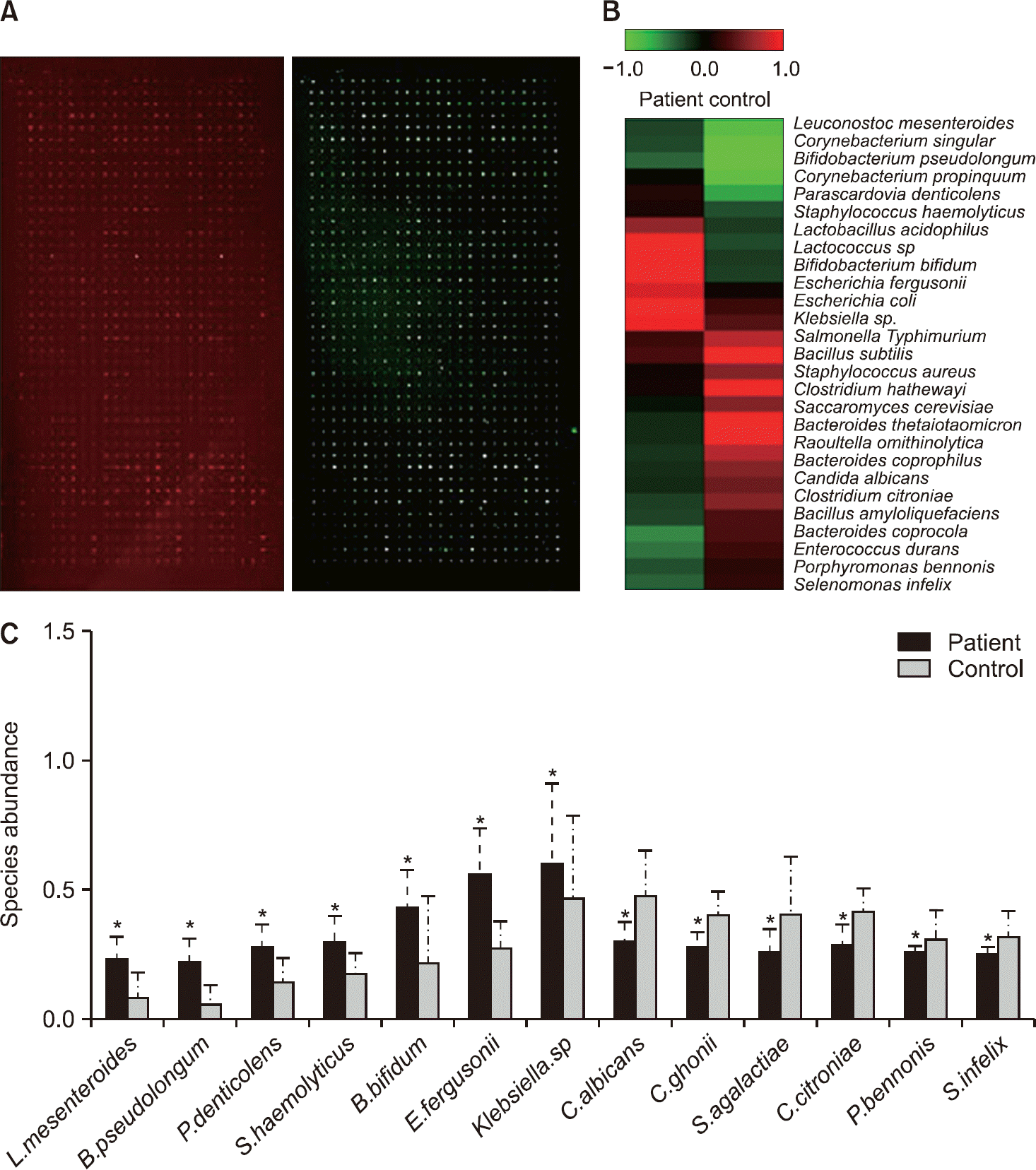 | Figure 2.Detection of disease-specific bacterial species using bacterial antibody microarray. (A) Representative images of gut bacterial antibody microarray in serum. (B) Differential expression at the bacterial species level between healthy controls and patients. (C) Species abundance analysis between healthy controls (n=11) and patients (n=8). Green and red col-ors indicate high and low expression of bacterial antibodies, respectively. *p<0.01. |
Table 1.
The alterations of gastrointestinal microbial composition in SSc patients
Table 1.
Continued




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download