Abstract
Viperin is an IFN-stimulated gene (ISG)-encoded protein that was identified in human primary macrophages treated with IFN-γ and in human primary fibroblasts infected with cytomegalovirus (CMV). This protein plays multiple roles in various cell types. It inhibits viral replication, mediates signaling pathways, and regulates cellular metabolism. Recent studies have shown that viperin inhibits IFN expression in macrophages, while it enhances TLR7 and TLR9-mediated IFN production in plasmacytoid dendritic cells, suggesting that viperin can play different roles in activation of the same pathway in different cell types. Viperin also controls induction of ISGs in macrophages. However, the effect of viperin on induction of ISGs in cell types other than macrophages is unknown. Here, we show that viperin differentially induces ISGs in 2 distinct cell types, macrophages and fibroblasts isolated from wild type and viperin knockout mice. Unlike in bone marrow-derived macrophages (BMDMs), viperin downregulates the expression levels of ISGs such as bone marrow stromal cell antigen-2, Isg15, Isg54, myxovirus resistance dynamin like GTPase 2, and guanylate binding protein 2 in murine embryonic fibroblasts (MEFs) treated with type I or II IFN. However, viperin upregulates expression of these ISGs in both BMDMs and MEFs stimulated with polyinosinic-polycytidylic acid or CpG DNA and infected with murine CMV. The efficiency of viral entry is inversely proportional to the expression levels of ISGs in both cell types. The data indicate that viperin differentially regulates induction of ISGs in a cell type-dependent manner, which might provide different innate immune responses in distinct cell types against infections.
Go to : 
References
1. Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008; 8:911–922.

2. de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001; 69:912–920.
3. Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014; 32:513–545.

4. Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018; 233:6425–6440.

5. Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010; 32:593–604.

6. Liu YC, Zou XB, Chai YF, Yao YM. Macrophage polarization in inflammatory diseases. Int J Biol Sci. 2014; 10:520–529.

7. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013; 496:445–455.

8. Mayer-Barber KD, Yan B. Clash of the Cytokine Titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell Mol Immunol. 2017; 14:22–35.

9. Kalliolias GD, Ivashkiv LB. Overview of the biology of type I interferons. Arthritis Res Ther. 2010; 12(Suppl 1):S1.

10. John SP, Sun J, Carlson RJ, Cao B, Bradfield CJ, Song J, Smelkinson M, Fraser ID. IFIT1 exerts opposing regulatory effects on the inflammatory and interferon gene programs in LPS-activated human macrophages. Cell Reports. 2018; 25:95–106. e6.

11. Tecalco Cruz AC, Mejía-Barreto K. Cell type-dependent regulation of free ISG15 levels and ISGylation. J Cell Commun Signal. 2017; 11:127–135.

12. Fan JB, Miyauchi-Ishida S, Arimoto K, Liu D, Yan M, Liu CW, Győrffy B, Zhang DE. Type I IFN induces protein ISGylation to enhance cytokine expression and augments colonic inflammation. Proc Natl Acad Sci U S A. 2015; 112:14313–14318.

13. Pang X, Li X, Mo Z, Huang J, Deng H, Lei Z, Zheng X, Feng Z, Xie D, Gao Z. IFI16 is involved in HBV-associated acute-on-chronic liver failure inflammation. BMC Gastroenterol. 2018; 18:61.

14. Trahey M, Weissman IL. Cyclophilin C-associated protein: a normal secreted glycoprotein that down-modulates endotoxin and proinflammatory responses in vivo. Proc Natl Acad Sci U S A. 1999; 96:3006–3011.

15. Lynch MD, Watt FM. Fibroblast heterogeneity: implications for human disease. J Clin Invest. 2018; 128:26–35.

16. Hamada A, Torre C, Drancourt M, Ghigo E. Trained immunity carried by non-immune cells. Front Microbiol. 2019; 9:3225.

17. Buechler MB, Turley SJ. A short field guide to fibroblast function in immunity. Semin Immunol. 2018; 35:48–58.

18. Jones PH, Mehta HV, Maric M, Roller RJ, Okeoma CM. Bone marrow stromal cell antigen 2 (BST-2) restricts mouse mammary tumor virus (MMTV) replication in vivo. Retrovirology. 2012; 9:10.
19. Perwitasari O, Cho H, Diamond MS, Gale M Jr. Inhibitor of κ B kinase ε (IKKε), STAT1, and IFIT2 proteins define novel innate immune effector pathway against West Nile virus infection. J Biol Chem. 2011; 286:44412–44423.

20. Tallóczy Z, Virgin HW 4th, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006; 2:24–29.

21. Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci U S A. 2001; 98:15125–15130.

22. Zhu H, Cong JP, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci U S A. 1997; 94:13985–13990.

23. Seo JY, Yaneva R, Cresswell P. Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe. 2011; 10:534–539.

24. Seo JY, Yaneva R, Hinson ER, Cresswell P. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science. 2011; 332:1093–1097.

25. Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008; 82:1665–1678.

26. Rivieccio MA, Suh HS, Zhao Y, Zhao ML, Chin KC, Lee SC, Brosnan CF. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J Immunol. 2006; 177:4735–4741.

27. Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007; 2:96–105.

28. Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol. 2007; 81:11246–11255.

29. Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T, Akira S. Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity. 2011; 34:352–363.

30. Eom J, Kim JJ, Yoon SG, Jeong H, Son S, Lee JB, Yoo J, Seo HJ, Cho Y, Kim KS, et al. Intrinsic expression of viperin regulates thermogenesis in adipose tissues. Proc Natl Acad Sci U S A. 2019; 116:17419–17428.

31. Qiu LQ, Cresswell P, Chin KC. Viperin is required for optimal Th2 responses and T-cell receptor-mediated activation of NF-κ B and AP-1. Blood. 2009; 113:3520–3529.

32. Seo JY, Cresswell P. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 2013; 9:e1003497.

33. Jang JS, Lee JH, Jung NC, Choi SY, Park SY, Yoo JY, Song JY, Seo HG, Lee HS, Lim DS. Rsad2 is necessary for mouse dendritic cell maturation via the IRF7-mediated signaling pathway. Cell Death Dis. 2018; 9:823.

34. Eom J, Yoo J, Kim JJ, Lee JB, Choi W, Park CG, Seo JY. Viperin deficiency promotes polarization of macrophages and secretion of M1 and M2 cytokines. Immune Netw. 2018; 18:e32.

35. Carissimo G, Teo TH, Chan YH, Lee CY, Lee B, Torres-Ruesta A, Tan JJ, Chua TK, Fong SW, Lum FM, et al. Viperin controls chikungunya virus-specific pathogenic T cell IFNγ Th1 stimulation in mice. Life Sci Alliance. 2019; 2:e201900298.
36. Hee JS, Cresswell P. Viperin interaction with mitochondrial antiviral signaling protein (MAVS) limits viperin-mediated inhibition of the interferon response in macrophages. PLoS One. 2017; 12:e0172236.

37. Ryu SH, Na HY, Sohn M, Han SM, Choi W, In H, Hong S, Jeon H, Seo JY, Ahn J, et al. Reduced expression of granule proteins during extended survival of eosinophils in splenocyte culture with GM-CSF. Immunol Lett. 2016; 173:7–20.

38. Lissner MM, Thomas BJ, Wee K, Tong AJ, Kollmann TR, Smale ST. Age-related gene expression differences in monocytes from human neonates, young adults, and older adults. PLoS One. 2015; 10:e0132061.

39. Josset L, Tchitchek N, Gralinski LE, Ferris MT, Eisfeld AJ, Green RR, Thomas MJ, Tisoncik-Go J, Schroth GP, Kawaoka Y, et al. Annotation of long non-coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014; 11:875–890.

40. Ouda R, Sarai N, Nehru V, Patel MC, Debrosse M, Bachu M, Chereji RV, Eriksson PR, Clark DJ, Ozato K. SPT6 interacts with NSD2 and facilitates interferon-induced transcription. FEBS Lett. 2018; 592:1681–1692.

41. Bachu M, Tamura T, Chen C, Narain A, Nehru V, Sarai N, Ghosh SB, Ghosh A, Kavarthapu R, Dufau ML, et al. A versatile mouse model of epitope-tagged histone H3.3 to study epigenome dynamics. J Biol Chem. 2019; 294:1904–1914.

42. Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005; 5:375–386.

43. Lindqvist R, Kurhade C, Gilthorpe JD, Överby AK. Cell-type- and region-specific restriction of neurotropic flavivirus infection by viperin. J Neuroinflammation. 2018; 15:80.

44. Platanitis E, Demiroz D, Schneller A, Fischer K, Capelle C, Hartl M, Gossenreiter T, Müller M, Novatchkova M, Decker T. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat Commun. 2019; 10:2921.

Go to : 
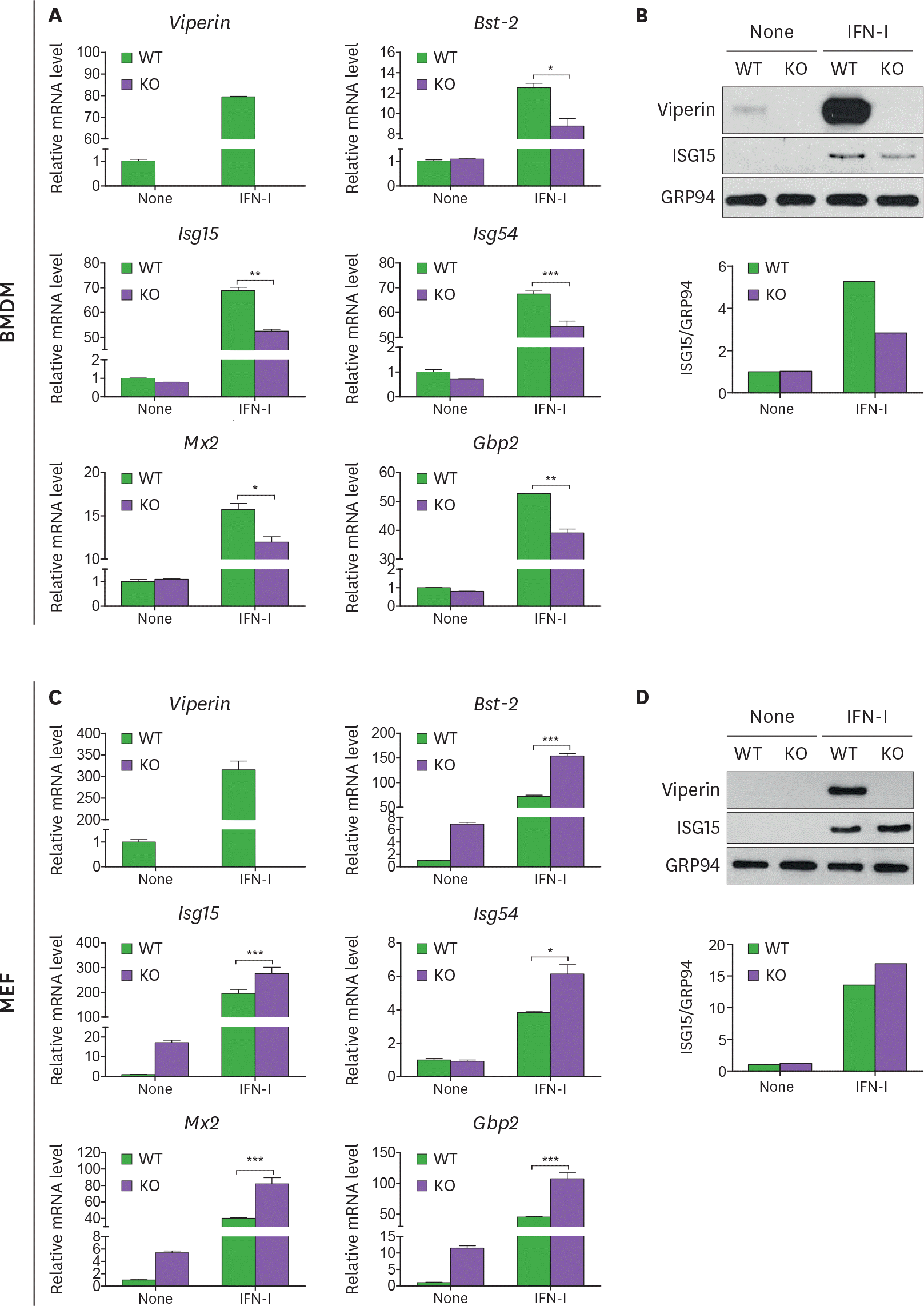 | Figure 1.Viperin differentially regulates ISG expression in BMDMs and MEFs treated with type I IFN. (A-D) The effect of viperin on expression of ISGs in BMDMs and MEFs upon type I IFN treatment. The bone marrow cells isolated from WT and viperin KO C57BL/6 mice were differentiated into BMDMs in M-CSF conditioned media for 7 days. The BMDMs were washed and plated in media without M-CSF for 24 h (A, B). MEFs were isolated from these mice and immortalized (C, D). The cells were treated with or without type I IFN (1,000 U/ml) for 24 h. The mRNA expression levels of ISGs in the cells were measured by qRT-PCR and normalized to β-actin mRNA (A, C). Data are presented as mean ± SEM of triplicate samples and are representative of three individual experiments. Expression of viperin and ISG15 proteins in the cells was detected by immunoblot using anti-viperin (MaP.VIP) or anti-ISG15 antibody (B, D). GRP94 served as a protein-loading control. Quantitation of ISG15 protein level was normalized to GRP94. * p<0.05; ** p<0.01; *** p<0.001. |
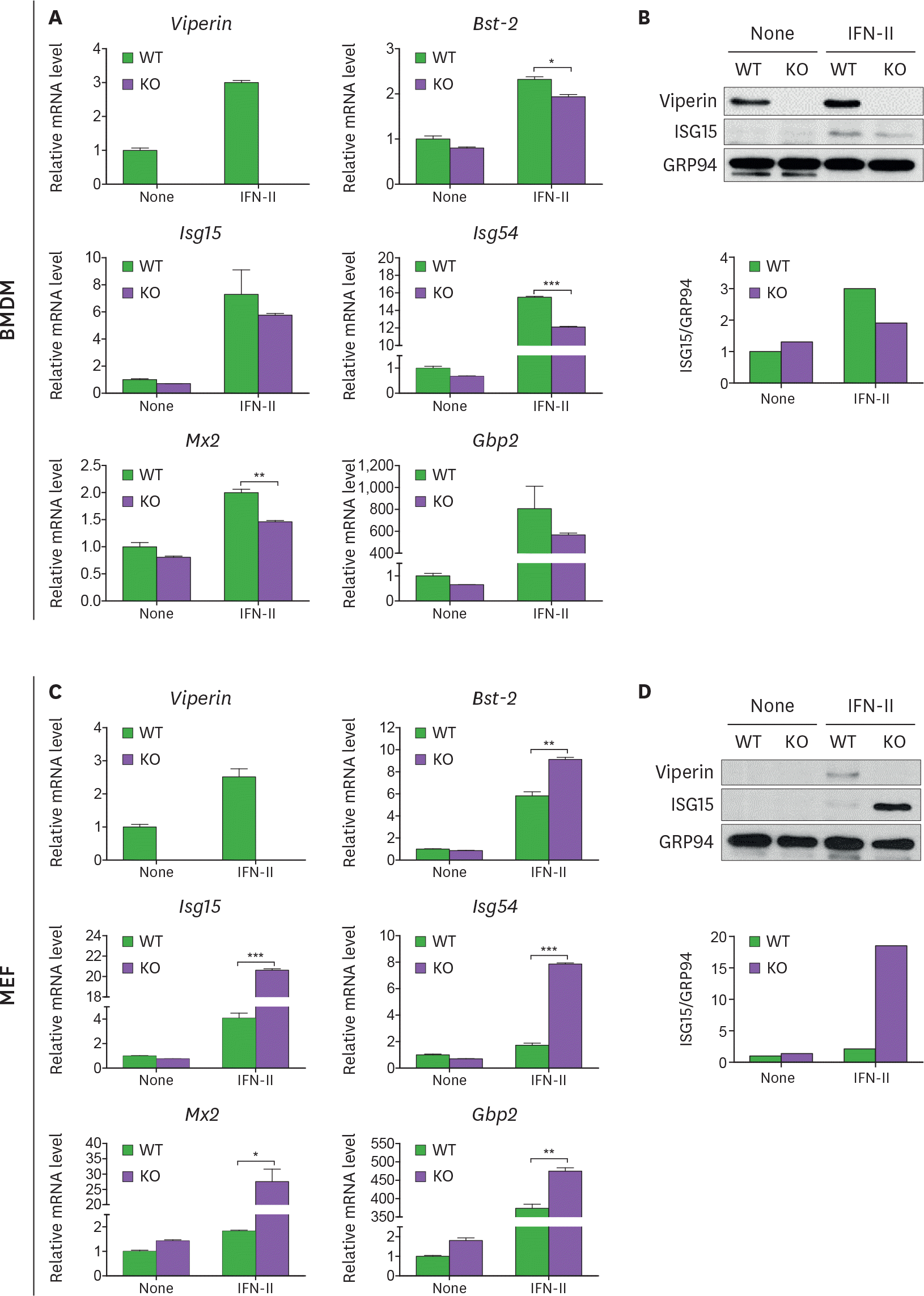 | Figure 2.Viperin differently induces ISG expression in BMDMs and MEFs treated with type II IFN. (A-D) The effect of viperin on expression of ISGs in BMDMs and MEFs upon type II IFN treatment. WT and viperin KO BMDMs (A, B) or MEFs (C, D) were treated with or without IFN-γ (100 ng/ml) for 24 h. The mRNA expression levels of ISGs in the cells were measured by qRT-PCR and normalized to β-actin mRNA (A, C). Data are presented as mean±SEM of triplicate samples and are representative of three individual experiments. Expression of viperin and ISG15 proteins in the cells was detected by immunoblot using anti-viperin (MaP.VIP) or anti-ISG15 antibody (B, D). GRP94 served as a protein-loading control. Quantitation of ISG15 protein level was normalized to GRP94. * p<0.05; ** p<0.01; *** p<0.001. |
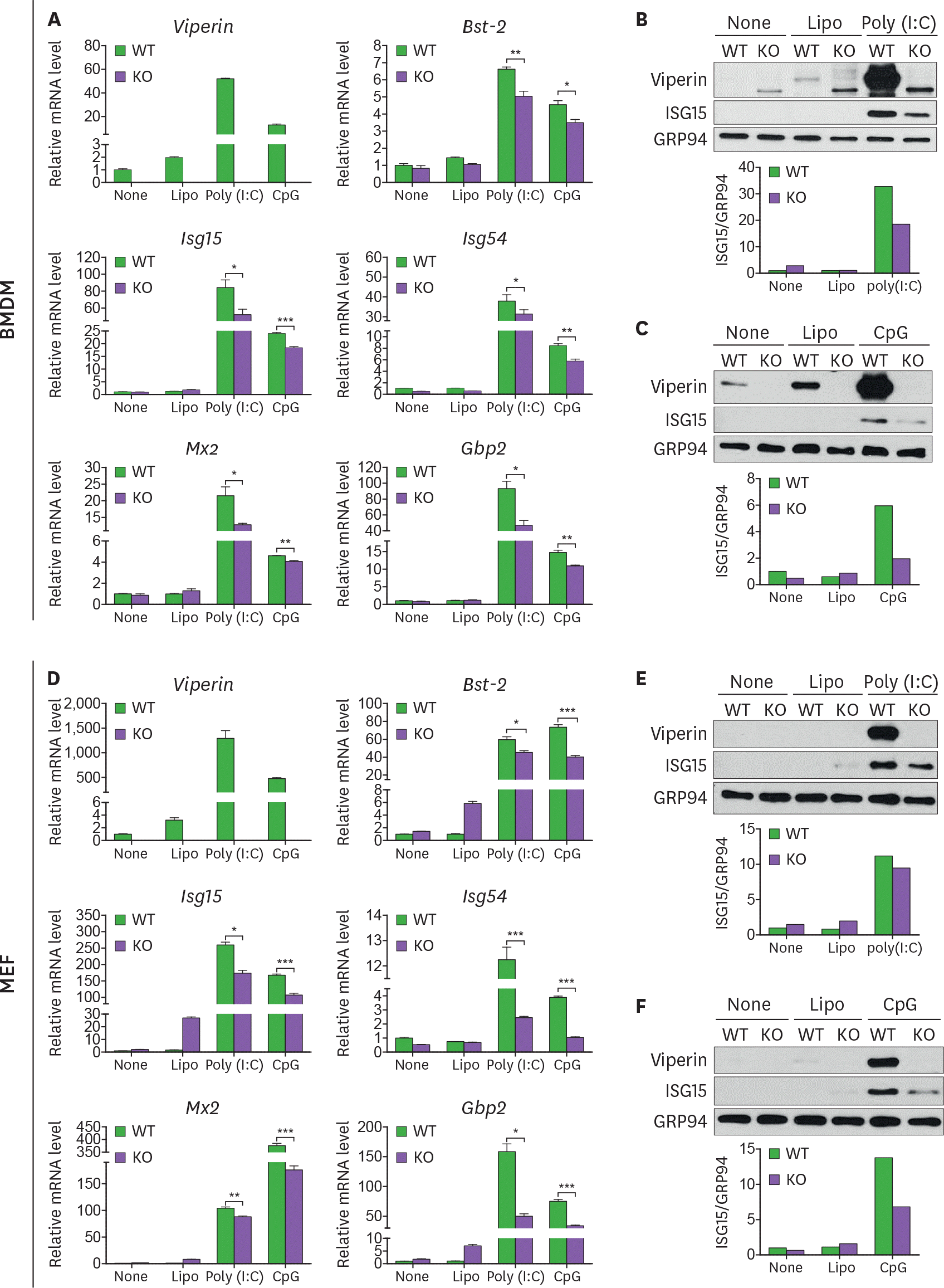 | Figure 3.Viperin enhances ISG expression in both BMDMs and MEFs transfected with poly(I:C) or CpG DNA. (A-F) The effect of viperin on expression of ISGs in BMDMs and MEFs upon poly(I:C) or CpG DNA treatment. WT and viperin KO BMDMs (A-C) or MEFs (D-F) were treated with lipofectamine 2000 (Lipo), or transfected with poly(I:C) (1 μg/ml) or CpG DNA (1 μg/ml) for 24 h. The mRNA expression levels of ISGs in the cells were measured by qRT-PCR and normalized to β-actin mRNA (A, D). Data are presented as mean±SEM of triplicate samples and are representative of three individual experiments. Expression of viperin and ISG15 proteins in the cells was detected by immunoblot using anti-viperin (MaP.VIP) or anti-ISG15 antibody (B, C, E, F). GRP94 served as a protein-loading control. Quantitation of ISG15 protein level was normalized to GRP94. * p<0.05; ** p<0.01; *** p<0.001. |
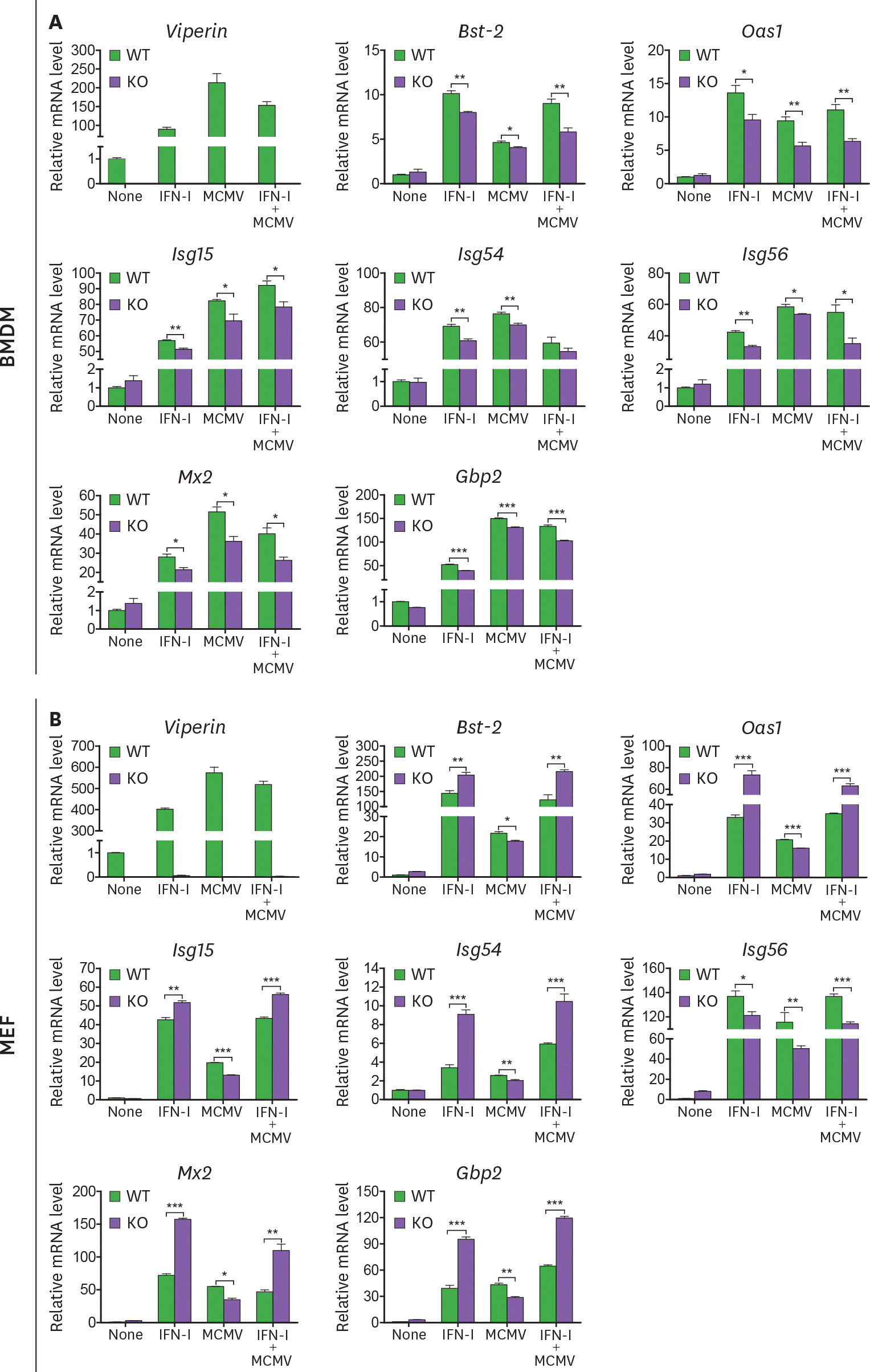 | Figure 4.Viperin differentially regulates ISG expression in BMDMs and MEFs treated with MCMV and/or type I IFN. (A, B) The effect of viperin on expression of ISGs in BMDMs and MEFs infected with MCMV in the absence or presence of type I IFN. WT and viperin KO BMDMs (A) or MEFs (B) were treated with or without type I IFN (1,000 U/ml) for 8 h and then infected with MCMV at an MOI of 0.2 for 24 h. The mRNA expression levels of ISGs in the cells were measured by qRT-PCR and normalized to β-actin mRNA. Data are presented as mean ± SEM of triplicate samples and are representative of three individual experiments. * p<0.05; ** p<0.01; *** p<0.001. |
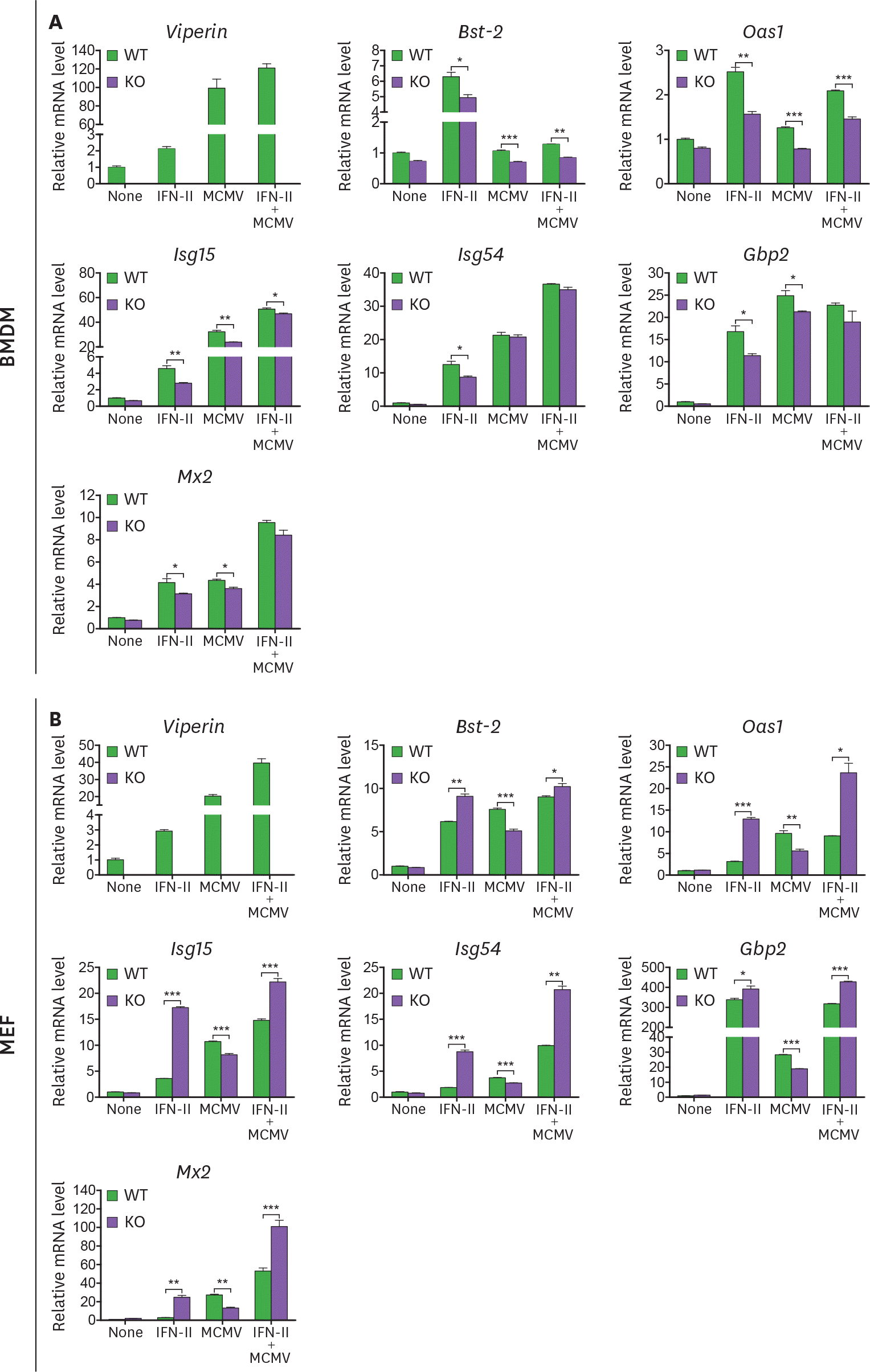 | Figure 5.Viperin differently induces ISG expression in BMDMs and MEFs treated with MCMV and/or type II IFN. (A, B) The effect of viperin on expression of ISGs in BMDMs and MEFs infected with MCMV in the absence or presence of type II IFN. WT and viperin KO BMDMs (A) or MEFs (B) were treated with or without IFN-γ (100 ng/ml) for 8 h and then infected with MCMV at an MOI of 0.2 for 24 h. The mRNA expression levels of ISGs in the cells were measured by qRT-PCR and normalized to β-actin mRNA. Data are presented as mean±SEM of triplicate samples and are representative of three individual experiments. * p<0.05; ** p<0.01; *** p<0.001. |
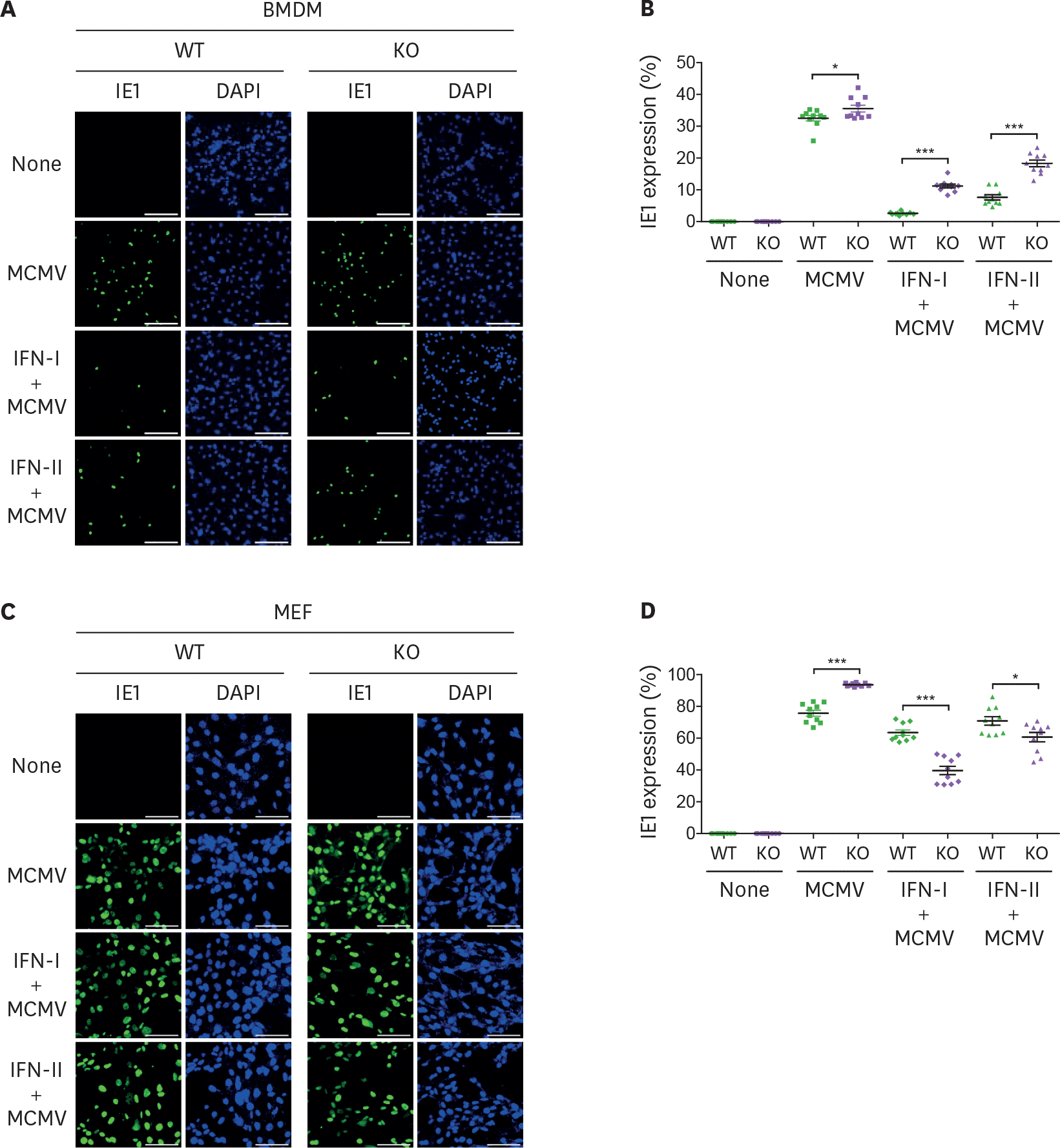 | Figure 6.MCMV entry is inversely proportional to viperin-mediated ISG expression. (A-D) The efficiency of MCMV entry into BMDMs and MEFs infected with MCMV in the absence or presence of IFNs. WT and viperin KO BMDMs (A, B) or MEFs (C, D) were treated with or without type I IFN (1,000 U/ml) or IFN-γ (100 ng/ml) for 8 h and then infected with MCMV at an MOI of 2 for 24 h. The cells were stained with antibody specific to the MCMV protein IE1 (green) to identify infected cells. Nuclei were stained with DAPI (blue). A representative image from two individual experiments was shown (scale bar=100 μm) (A, C). The efficiency of MCMV entry into the cells was quantitated (B, D). The infected cells were counted in each image (n=10). The percentage of MCMV positive cells per total cells in each image was calculated. Data are presented as mean±SEM. * p<0.05; *** p<0.001. |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download