Abstract
Our current knowledge of mycobacterial infections in humans has progressively increased over the past few decades. The infection of Mycobacterium tuberculosis causes tuberculosis (TB) disease, which has reasoned for excessive morbidity and mortality worldwide, and has become a foremost issue of health problem globally. Mycobacterium leprae, another member of the family Mycobacteriaceae, is responsible for causing a chronic disease known as leprosy that mainly affects mucosa of the upper respiratory tract, skin, peripheral nerves, and eyes. Ample amount of existing data suggests that pathogenic mycobacteria have skilled in utilizing different mechanisms to escape or offset the host immune responses. They hijack the machinery of immune cells through the modulation of microRNAs (miRs), which regulate gene expression and immune responses of the host. Evidence shows that miRs have now gained considerable attention in the research, owing to their involvement in a broad range of inflammatory processes that are further implicated in the pathogenesis of several diseases. However, the knowledge of functions of miRs during mycobacterial infections remains limited. This review summarises recent findings of differential expression of miRs, which are used to good advantage by mycobacteria in offsetting host immune responses generated against them.
Go to : 
References
1. Gutierrez MC, Brisse S, Brosch R, Fabre M, Omaïs B, Marmiesse M, Supply P, Vincent V. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005; 1:e5.
2. World Health Organization (WHO). Global tuberculosis report 2018 [Internet]. Available at. https://www.who.int/tb/publications/global_report/en/. [accessed on 1 April 2019].
3. Fischer M. Leprosy – an overview of clinical features, diagnosis, and treatment. J Dtsch Dermatol Ges. 2017; 15:801–827.

4. World Health Organization (WHO). WHO fact sheets: leprosy [Internet]. Available at. https://www.who.int/news-room/fact-sheets/detail/leprosy. [accessed on 12 August 2019].
5. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007; 175:367–416.

6. Parent LJ, Salam MM, Appelbaum PC, Dossett JH. Disseminated Mycobacterium marinum infection and bacteremia in a child with severe combined immunodeficiency. Clin Infect Dis. 1995; 21:1325–1327.

7. Watanabe Y, Kanai A. Systems biology reveals microRNA-mediated gene regulation. Front Genet. 2011; 2:29.

8. Sabir N, Hussain T, Shah SZ, Peramo A, Zhao D, Zhou X. miRNAs in Tuberculosis: New avenues for diagnosis and host-directed therapy. Front Microbiol. 2018; 9:602.

9. Qin Y, Wang Q, Zhou Y, Duan Y, Gao Q. Inhibition of IFN-γ-induced nitric oxide dependent antimycobacterial activity by miR-155 and C/EBPβ. Int J Mol Sci. 2016; 17:535.

10. Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. 2011; 6:e20258.
11. Guirado E, Schlesinger LS, Kaplan G. Macrophages in tuberculosis: friend or foe. Semin Immunopathol. 2013; 35:563–583.

12. Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. 2009; 10:943–948.

13. Mack U, Migliori GB, Sester M, Rieder HL, Ehlers S, Goletti D, Bossink A, Magdorf K, Hölscher C, Kampmann B, et al. LTBI: latent tuberculosis infection or lasting immune responses to M. tuberculosis? A TBNET consensus statement. Eur Respir J. 2009; 33:956–973.

14. Lee JY. Diagnosis and treatment of extrapulmonary tuberculosis. Tuberc Respir Dis (Seoul). 2015; 78:47–55.

15. Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, Bloom BR. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995; 2:561–572.

16. Roach DR, Bean AG, Demangel C, France MP, Briscoe H, Britton WJ. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J Immunol. 2002; 168:4620–4627.

18. Chavarro-Portillo B, Soto CY, Guerrero MI. Mycobacterium leprae's evolution and environmental adaptation. Acta Trop. 2019; 197:105041.
19. Jin SH, An SK, Lee SB. The formation of lipid droplets favors intracellular Mycobacterium leprae survival in SW-10, non-myelinating Schwann cells. PLoS Negl Trop Dis. 2017; 11:e0005687.
20. Inkeles MS, Teles RM, Pouldar D, Andrade PR, Madigan CA, Lopez D, Ambrose M, Noursadeghi M, Sarno EN, Rea TH, et al. Cell-type deconvolution with immune pathways identifies gene networks of host defense and immunopathology in leprosy. JCI Insight. 2016; 1:e88843.

21. Lyrio EC, Campos-Souza IC, Corrêa LC, Lechuga GC, Verícimo M, Castro HC, Bourguignon SC, Côrte-Real S, Ratcliffe N, Declercq W, et al. Interaction of Mycobacterium leprae with the HaCaT human keratinocyte cell line: new frontiers in the cellular immunology of leprosy. Exp Dermatol. 2015; 24:536–542.

22. Falkinham JO 3rd. Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol. 2009; 107:356–367.

23. Falkinham JO. Impact of human activities on the ecology of nontuberculous mycobacteria. Future Microbiol. 2010; 5:951–960.

24. Williams MM, Yakrus MA, Arduino MJ, Cooksey RC, Crane CB, Banerjee SN, Hilborn ED, Donlan RM. Structural analysis of biofilm formation by rapidly and slowly growing nontuberculous mycobacteria. Appl Environ Microbiol. 2009; 75:2091–2098.

25. Orme IM, Ordway DJ. Host response to nontuberculous mycobacterial infections of current clinical importance. Infect Immun. 2014; 82:3516–3522.

26. Bermudez LE, Young LS. Tumor necrosis factor, alone or in combination with IL-2, but not IFN-gamma, is associated with macrophage killing of Mycobacterium avium complex. J Immunol. 1988; 140:3006–3013.
27. Tezera LB, Bielecka MK, Chancellor A, Reichmann MT, Shammari BA, Brace P, Batty A, Tocheva A, Jogai S, Marshall BG, et al. Dissection of the host-pathogen interaction in human tuberculosis using a bioengineered 3-dimensional model. eLife. 2017; 6:e21283.

28. O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010; 10:111–122.
29. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004; 432:231–235.

30. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018; 9:402.
31. Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009; 326:1275–1279.

32. Song MS, Rossi JJ. Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem J. 2017; 474:1603–1618.

33. Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008; 455:58–63.

35. Bettencourt P, Pires D, Anes E. Immunomodulating microRNAs of mycobacterial infections. Tuberculosis (Edinb). 2016; 97:1–7.

36. Singh PK, Singh AV, Chauhan DS. Current understanding on micro RNAs and its regulation in response to Mycobacterial infections. J Biomed Sci. 2013; 20:14.

37. Krysko DV, Vanden Berghe T, D'Herde K, Vandenabeele P. Apoptosis and necrosis: detection, discrimination and phagocytosis. Methods. 2008; 44:205–221.

38. Fratazzi C, Arbeit RD, Carini C, Balcewicz-Sablinska MK, Keane J, Kornfeld H, Remold HG. Macrophage apoptosis in mycobacterial infections. J Leukoc Biol. 1999; 66:763–764.

41. Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX, Divangahi M, Remold HG. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 2011; 4:279–287.

42. Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995; 82:495–506.

43. Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. RIPK1 is not essential for TNFR1-induced activation of NF-κ B. Cell Death Differ. 2010; 17:482–487.

44. Park YC, Ye H, Hsia C, Segal D, Rich RL, Liou HC, Myszka DG, Wu H. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell. 2000; 101:777–787.

45. Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011; 12:439–452.

46. Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008; 30:689–700.

47. Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003; 114:181–190.

48. de Almagro MC, Vucic D. The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti-cancer therapy. Exp Oncol. 2012; 34:200–211.
49. Waring P, Müllbacher A. Cell death induced by the Fas/Fas ligand pathway and its role in pathology. Immunol Cell Biol. 1999; 77:312–317.

50. Adrain C, Creagh EM, Martin SJ. Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2. EMBO J. 2001; 20:6627–6636.

51. Chinnaiyan AM. The apoptosome: heart and soul of the cell death machine. Neoplasia. 1999; 1:5–15.

52. Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ. Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J. 2004; 23:2134–2145.

53. Chauhan D, Li G, Hideshima T, Podar K, Mitsiades C, Mitsiades N, Munshi N, Kharbanda S, Anderson KC. JNK-dependent release of mitochondrial protein, Smac, during apoptosis in multiple myeloma (MM) cells. J Biol Chem. 2003; 278:17593–17596.

55. Deng Y, Ren X, Yang L, Lin Y, Wu X. A JNK-dependent pathway is required for TNFα-induced apoptosis. Cell. 2003; 115:61–70.

56. Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. 2017; 13:815–827.

57. Hagenbuchner J, Ausserlechner MJ. Mitochondria and FOXO3: breath or die. Front Physiol. 2013; 4:147.

58. Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, Suar M, Van Kaer L, Bishai WR, Das G. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem. 2013; 288:5056–5061.

59. Rajaram MV, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, Schoenberg DR, Torrelles JB, Schlesinger LS. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U S A. 2011; 108:17408–17413.

60. Tripathi A, Srivastava V, Singh BN. hsa-let-7b–5p facilitates Mycobacterium tuberculosis survival in THP-1 human macrophages by Fas downregulation. FEMS Microbiol Lett. 2018; 365:365.
61. Zhang G, Liu X, Wang W, Cai Y, Li S, Chen Q, Liao M, Zhang M, Zeng G, Zhou B, et al. Down-regulation of miR-20a–5p triggers cell apoptosis to facilitate mycobacterial clearance through targeting JNK2 in human macrophages. Cell Cycle. 2016; 15:2527–2538.

62. Liang S, Song Z, Wu Y, Gao Y, Gao M, Liu F, Wang F, Zhang Y. MicroRNA-27b modulates inflammatory response and apoptosis during Mycobacterium tuberculosis infection. J Immunol. 2018; 200:3506–3518.

63. Wang Q, Liu S, Tang Y, Liu Q, Yao Y. MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS One. 2014; 9:e100949.
64. Liu Y, Jiang J, Wang X, Zhai F, Cheng X. miR-582–5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PLoS One. 2013; 8:e78381.

65. Xi X, Zhang C, Han W, Zhao H, Zhang H, Jiao J. MicroRNA-223 is upregulated in active tuberculosis patients and inhibits apoptosis of macrophages by targeting FOXO3. Genet Test Mol Biomarkers. 2015; 19:650–656.

66. Zhang C, Xi X, Wang Q, Jiao J, Zhang L, Zhao H, Lai Z. The association between serum miR-155 and natural killer cells from tuberculosis patients. Int J Clin Exp Med. 2015; 8:9168–9172.
67. Huang J, Jiao J, Xu W, Zhao H, Zhang C, Shi Y, Xiao Z. MiR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol Med Rep. 2015; 12:7102–7108.

68. Kumar R, Halder P, Sahu SK, Kumar M, Kumari M, Jana K, Ghosh Z, Sharma P, Kundu M, Basu J. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell Microbiol. 2012; 14:1620–1631.

69. Tsai MC, Chakravarty S, Zhu G, Xu J, Tanaka K, Koch C, Tufariello J, Flynn J, Chan J. Characterization of the tuberculous granuloma in murine and human lungs: cellular composition and relative tissue oxygen tension. Cell Microbiol. 2006; 8:218–232.

70. Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev. 2015; 264:182–203.

72. Hoppe AD, Swanson JA. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol Biol Cell. 2004; 15:3509–3519.

73. Jongstra-Bilen J, Harrison R, Grinstein S. Fcγ-receptors induce Mac-1 (CD11b/CD18) mobilization and accumulation in the phagocytic cup for optimal phagocytosis. J Biol Chem. 2003; 278:45720–45729.

74. Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996; 15:5326–5335.

75. Miki H, Takenawa T. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem Biophys Res Commun. 1998; 243:73–78.

76. Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999; 97:221–231.

77. Ma L, Rohatgi R, Kirschner MW. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc Natl Acad Sci U S A. 1998; 95:15362–15367.

78. Doherty TM, Arditi M. TB, or not TB: that is the question – does TLR signaling hold the answer? J Clin Invest. 2004; 114:1699–1703.
79. Jo EK, Yang CS, Choi CH, Harding CV. Intracellular signalling cascades regulating innate immune responses to Mycobacteria: branching out from Toll-like receptors. Cell Microbiol. 2007; 9:1087–1098.

80. Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol. 2011; 2011:405310.
81. Basu J, Shin DM, Jo EK. Mycobacterial signaling through Toll-like receptors. Front Cell Infect Microbiol. 2012; 2:145.

82. Deguine J, Barton GM. MyD88: a central player in innate immune signaling. F1000Prime Rep. 2014; 6:97.

83. Zhang J, Clark K, Lawrence T, Peggie MW, Cohen P. An unexpected twist to the activation of IKKβ: TAK1 primes IKKβ for activation by autophosphorylation. Biochem J. 2014; 461:531–537.

85. Harris J, Hope JC, Keane J. Tumor necrosis factor blockers influence macrophage responses to Mycobacterium tuberculosis. J Infect Dis. 2008; 198:1842–1850.

86. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008; 8:958–969.

87. Beg AA, Baltimore D. An essential role for NF-κ B in preventing TNF-α-induced cell death. Science. 1996; 274:782–784.

88. Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-α-induced apoptosis by NF-κ B. Science. 1996; 274:787–789.
89. Keane J, Balcewicz-Sablinska MK, Remold HG, Chupp GL, Meek BB, Fenton MJ, Kornfeld H. Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect Immun. 1997; 65:298–304.

90. Bettencourt P, Marion S, Pires D, Santos LF, Lastrucci C, Carmo N, Blake J, Benes V, Griffiths G, Neyrolles O, et al. Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142–3p. Front Cell Infect Microbiol. 2013; 3:19.

91. Xu G, Zhang Z, Wei J, Zhang Y, Zhang Y, Guo L, Liu X. microR-142–3p down-regulates IRAK-1 in response to Mycobacterium bovis BCG infection in macrophages. Tuberculosis (Edinb). 2013; 93:606–611.

92. Wang J, Yang K, Zhou L, Minhaowu , Wu Y, Zhu M, Lai X, Chen T, Feng L, Li M, et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 2013; 9:e1003697.

93. Etna MP, Sinigaglia A, Grassi A, Giacomini E, Romagnoli A, Pardini M, Severa M, Cruciani M, Rizzo F, Anastasiadou E, et al. Mycobacterium tuberculosis-induced miR-155 subverts autophagy by targeting ATG3 in human dendritic cells. PLoS Pathog. 2018; 14:e1006790.
94. Wang J, Zhu X, Xiong X, Ge P, Liu H, Ren N, Khan FA, Zhou X, Zhang L, Yuan X, et al. Identification of potential urine proteins and microRNA biomarkers for the diagnosis of pulmonary tuberculosis patients. Emerg Microbes Infect. 2018; 7:63.

95. Iwai H, Funatogawa K, Matsumura K, Kato-Miyazawa M, Kirikae F, Kiga K, Sasakawa C, Miyoshi-Akiyama T, Kirikae T. MicroRNA-155 knockout mice are susceptible to Mycobacterium tuberculosis infection. Tuberculosis (Edinb). 2015; 95:246–250.

96. Ma C, Li Y, Li M, Deng G, Wu X, Zeng J, Hao X, Wang X, Liu J, Cho WC, et al. microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol Immunol. 2014; 62:150–158.

97. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κ B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006; 103:12481–12486.

98. Nathan C. Inducible nitric oxide synthase in the tuberculous human lung. Am J Respir Crit Care Med. 2002; 166:130–131.

99. Rangel Moreno J, Estrada García I, De La Luz García Hernández M, Aguilar Leon D, Marquez R, Hernández Pando R. The role of prostaglandin E2 in the immunopathogenesis of experimental pulmonary tuberculosis. Immunology. 2002; 106:257–266.

100. Liu Z, Zhou G, Deng X, Yu Q, Hu Y, Sun H, Wang Z, Chen H, Jia C, Wang D. Analysis of miRNA expression profiling in human macrophages responding to Mycobacterium infection: induction of the immune regulator miR-146a. J Infect. 2014; 68:553–561.

101. Wei J, Huang X, Zhang Z, Jia W, Zhao Z, Zhang Y, Liu X, Xu G. MyD88 as a target of microRNA-203 in regulation of lipopolysaccharide or Bacille Calmette-Guerin induced inflammatory response of macrophage RAW264.7 cells. Mol Immunol. 2013; 55:303–309.

102. Ghorpade DS, Holla S, Kaveri SV, Bayry J, Patil SA, Balaji KN. Sonic hedgehog-dependent induction of microRNA 31 and microRNA 150 regulates Mycobacterium bovis BCG-driven toll-like receptor 2 signaling. Mol Cell Biol. 2013; 33:543–556.

103. Xu G, Zhang Z, Xing Y, Wei J, Ge Z, Liu X, Zhang Y, Huang X. MicroRNA-149 negatively regulates TLR-triggered inflammatory response in macrophages by targeting MyD88. J Cell Biochem. 2014; 115:919–927.

104. von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008; 321:691–696.

105. Wu Z, Lu H, Sheng J, Li L. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett. 2012; 586:2459–2467.

106. Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, Gupta P, Raffetseder J, Lerm M, Ghosh Z, et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κ B pathway. Cell Host Microbe. 2015; 17:345–356.

107. Liu Y, Wang R, Jiang J, Yang B, Cao Z, Cheng X. miR-223 is upregulated in monocytes from patients with tuberculosis and regulates function of monocyte-derived macrophages. Mol Immunol. 2015; 67:475–481.

108. Dorhoi A, Iannaccone M, Farinacci M, Faé KC, Schreiber J, Moura-Alves P, Nouailles G, Mollenkopf HJ, Oberbeck-Müller D, Jörg S, et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest. 2013; 123:4836–4848.

109. Keane J, Shurtleff B, Kornfeld H. TNF-dependent BALB/c murine macrophage apoptosis following Mycobacterium tuberculosis infection inhibits bacillary growth in an IFN-γ independent manner. Tuberculosis (Edinb). 2002; 82:55–61.

110. Larsen L, Röpke C. Suppressors of cytokine signalling: SOCS. APMIS. 2002; 110:833–844.
111. Mühl H, Pfeilschifter J. Anti-inflammatory properties of pro-inflammatory interferon-γ. Int Immunopharmacol. 2003; 3:1247–1255.

112. Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993; 178:2249–2254.

113. Lu YJ, Shen N, Wang X. Genetic associations between miR-146a/499 polymorphisms and tuberculosis: a meta-analysis. Int J Clin Exp Med. 2016; 9:6445–6452.
114. Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011; 12:861–869.

115. Ni B, Rajaram MV, Lafuse WP, Landes MB, Schlesinger LS. Mycobacterium tuberculosis decreases human macrophage IFN-γ responsiveness through miR-132 and miR-26a. J Immunol. 2014; 193:4537–4547.

116. Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol. 2011; 48:1084–1090.

117. Shi G, Mao G, Xie K, Wu D, Wang W. MiR-1178 regulates mycobacterial survival and inflammatory responses in Mycobacterium tuberculosis-infected macrophages partly via TLR4. J Cell Biochem. 2018; 119:7449–7457.

118. Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, Mehta MD, Vazirnia A, Rea TH, Sarno EN, et al. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat Med. 2012; 18:267–273.

120. Rivas-Santiago B, Schwander SK, Sarabia C, Diamond G, Klein-Patel ME, Hernandez-Pando R, Ellner JJ, Sada E. Human β-defensin 2 is expressed and associated with Mycobacterium tuberculosis during infection of human alveolar epithelial cells. Infect Immun. 2005; 73:4505–4511.

121. Mangalam AK, Taneja V, David CS. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol. 2013; 190:513–518.

122. Smart M, Behrens M, David L, Conway C, Taneja V. Immune response to immunodominant Mycobacterium tuberculosis antigen ESAT-6 derived peptide is HLA-haplotype dependent. Jacobs J Allergy Immunol. 2014; 1:002.
123. Costantino CM, Ploegh HL, Hafler DA. Cathepsin S regulates class II MHC processing in human CD4+ HLA-DR+ T cells. J Immunol. 2009; 183:945–952.

124. Driessen C, Bryant RA, Lennon-Duménil AM, Villadangos JA, Bryant PW, Shi GP, Chapman HA, Ploegh HL. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. J Cell Biol. 1999; 147:775–790.

125. Pires D, Bernard EM, Pombo JP, Carmo N, Fialho C, Gutierrez MG, Bettencourt P, Anes E. Mycobacterium tuberculosis modulates miR-106b–5p to control cathepsin S expression resulting in higher pathogen survival and poor T-cell activation. Front Immunol. 2017; 8:1819.
Go to : 
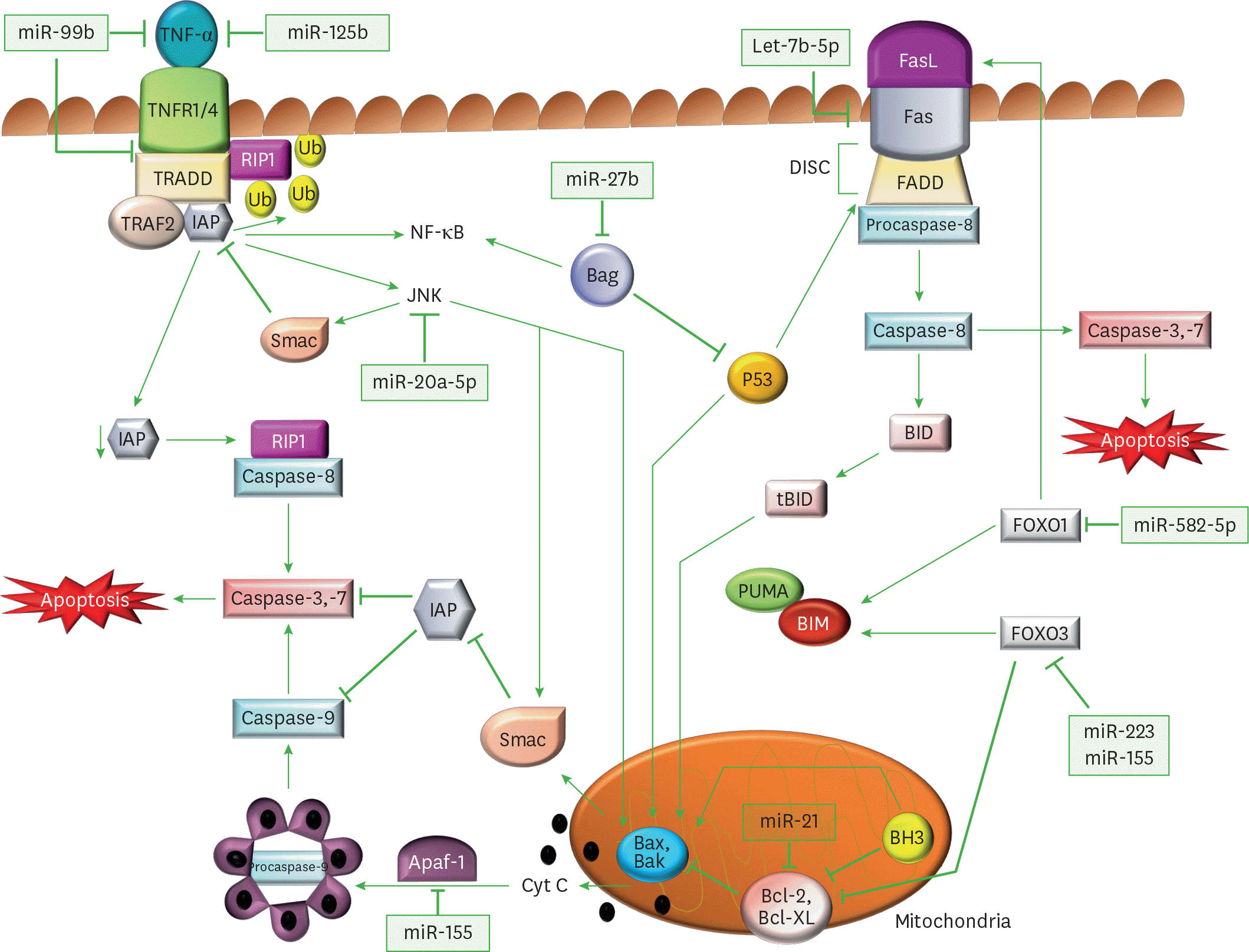 | Figure 1.Regulation of apoptotic signaling pathways through miRs by M. tuberculosis. Diagrammatic representation of miRs and their target genes, which alter the canonical apoptotic pathways of immune cells, and assist in M. tuberculosis survival. In the extrinsic apoptotic pathway, TNFα binds to its receptors (TNFR1/4) and triggers its trimerization, which recruits TRADD, an adaptor molecule that allows binding of TRAF-2 and IAP to the receptor complex. RIP1, a kinase then binds TNFR1 through TRADD association. IAP proteins are responsible for RIP1 ubiquitination, and in their absence, RIP1 cannot be ubiquitinated. Non-ubiquitinated RIP1 can form a cytosolic complex with the caspase-8, leading to induction of apoptosis. The binding of TRAF-2 to the receptor complex stimulates the JNK pathway, which is a proapoptotic pathway. JNK can promote the release of Smac, from mitochondria, which inhibits TRAF2/IAP complex formation, and relieves the inhibition of caspase-8, thereby triggering caspase activation. JNK also participates in intrinsic apoptotic pathway, as after activation, JNK translocates to the mitochondria and phosphorylates BH3-only family of Bcl-2 proteins, which antagonize the anti-apoptotic activity of Bcl-2 or Bcl-Xl proteins. Further, JNK stimulates the release cyt C from mitochondrial inner membrane through BID-Bax dependent mechanism and promotes apoptosomes formation from released cyt C, caspase-9, and Apaf-1. The apoptosomes initiates caspase-9-dependent caspase cascade. In FasR pathway, a FasL binds to FasR and induces the recruitment of FADD followed by activation of procaspase-8, which further activates caspase-3 (the executioner) causing apoptosis. Caspase-8 also cleaves BID, a BH3 domain-containing protein of the BCL-2, which triggers intrinsic apoptosis and amplifies the signal from the extrinsic pathway. The expression of another member of BH3 only protein is increased by FOXO1 and FOXO3, which initiates the Bax/Bak-dependent apoptotic pathway. |
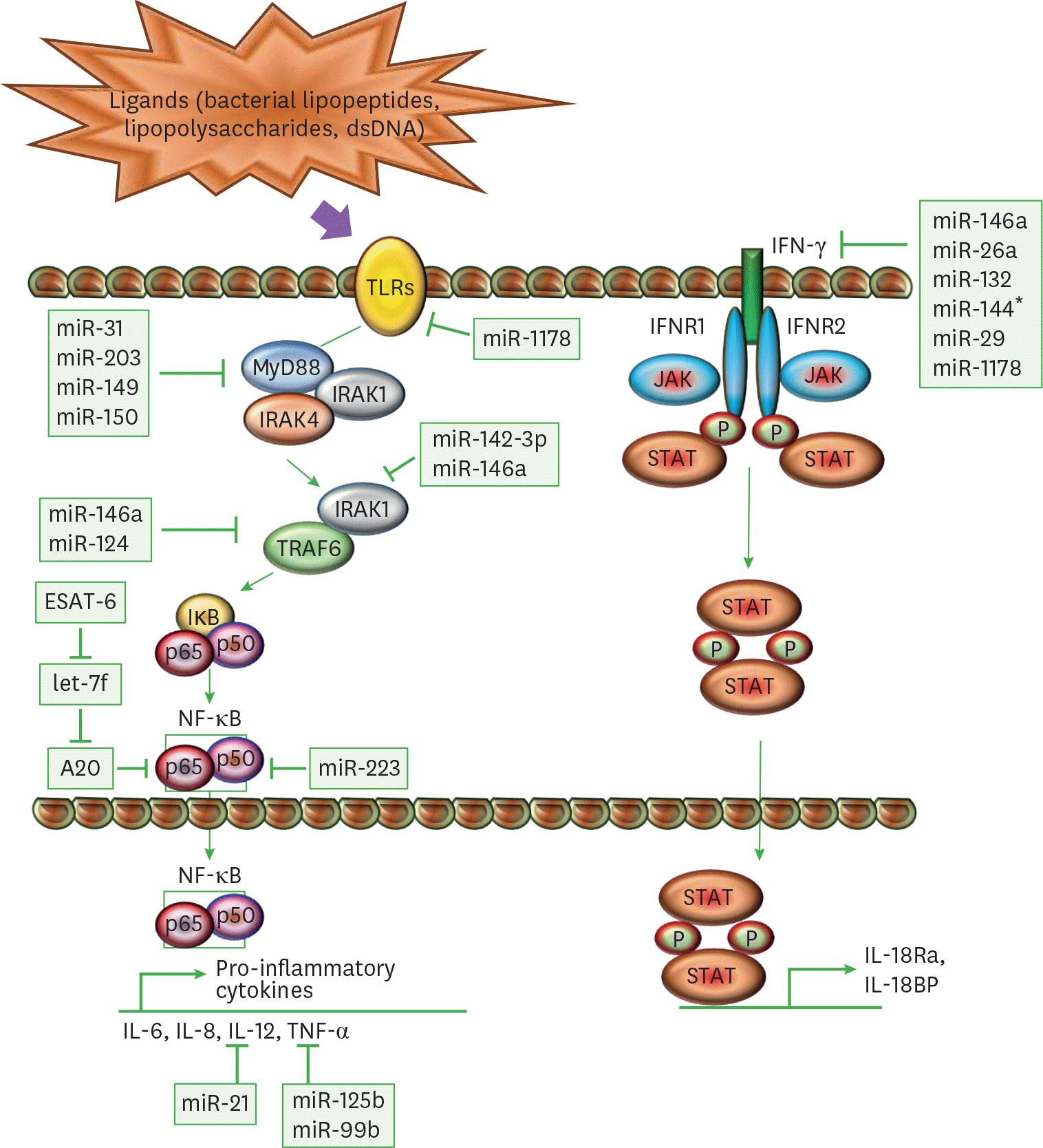 | Figure 2.Immunomodulation of macrophages through miRs. A schematic diagram represents different miRs and their target genes, which allows M. tuberculosis to mitigate the immune response through modulation of TLR and IFN-γ pathway. The ligation of mycobacterial ligands to TLRs leads to the interaction of MyD88 with IRAK4 and IRAK1. IRAK4 auto-phosphorylates and activates IRAK1. This promotes the activation of TRAF6, which further activates TAK1. TAK1 gives rise to canonical IKK complex by phosphorylating IKKα and IKKβ, which phosphorylates components of the transcription factor NF-κ B such as Iκ Bα, p50, and p65. TAK1-mediated activation of NF-κ B transcription factors drives the production of pro-inflammatory cytokines such as IL-12 and TNF-α. In another pathway, IFN-γ acts as a principal mediator of macrophage activation, which modulates pro-inflammatory cytokine production and induces production of anti-inflammatory molecules. Upon ligand binding, oligomerization and transphosphorylation of IFN-γ receptors (IFNR1 and IFNR2) activate JAK1 and JAK2, which creates a docking site for STAT1. STAT1 homodimerizes upon phosphorylation (P) in an antiparallel configuration, forming a complex γ-activated factor, which translocates to the nucleus and binds to the γ-activated site, located at the promoters of primary response genes, increasing their transcription, like IL-1Ra and IL-18BP. SOCS proteins negatively regulate the IFN-γ pathway by inhibiting JAKs and STAT1 phosphorylation. |
Table 1.
Regulation of host miRs during M. tuberculosis infections
Table 2.
miR-mediated modulation of host immune response during M. bovis and other mycobacterial infections




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download