Tuberculosis (TB), caused by members of the
Mycobacterium tuberculosis complex (MTBC), mainly
M. tuberculosis (MTB), is one of the top 10 causes of death worldwide [
1]. While MTB is an obligate human pathogen without an environmental reservoir, nontuberculous mycobacteria (NTM), which include mycobacteria other than MTBC and
Mycobacterium leprae , are ubiquitous organisms frequently isolated from soil and water [
23]. A recent epidemiological study of the incidence and prevalence of NTM infection identified rapid growth in the number of patients with NTM pulmonary disease (NTM-PD) in Korea, consistent with the global trend of increasing NTM infections [
4]. Owing to the increasing incidence and prevalence of both MTBC and NTM in Korea, rapid and accurate differentiation between MTBC and NTM infections is crucial for appropriate treatment and infection control [
5].
Detection of acid-fast bacilli (AFB) by microscopic examination is a rapid and inexpensive method for TB diagnosis; however, this approach does not differentiate between MTBC and NTM, which requires an additional phenotypic identification step using biochemical assays that are time-consuming and labor intensive [
67]. To overcome these disadvantages, various molecular diagnostic assays for differential identification of MTBC and NTM have been developed and evaluated [
89101112].
The GENEDIA MTB/NTM Detection Kit (GENEDIA MTB/NTM; Green Cross Medical Science Corp., Chungbuk, Korea) is a multiplex real-time PCR assay used for differential identification of MTBC and NTM. This assay targets the IS
6110 region of MTB, the internal transcribed spacer region, and the
rpoB gene of NTM. Previously, we evaluated the performance of this assay using liquid media [
8]. While the performance of GENEDIA MTB Detection Kit (GENEDIA MTB; Green Cross Medical Science Corp.) has been studied using respiratory specimens [
13], the performance of GENEDIA MTB/NTM with direct respiratory specimens has not been studied to date. We evaluated and compared the performance of GENEDIA MTB/NTM and the GENEDIA MTB assays using sputum specimens.
We used a total of 687 non-selective, consecutive sputum specimens submitted for AFB smear and mycobacterial culture between January and March 2018. This prospective study was approved by the Institutional Review Board of Samsung Medical Center, Seoul, Korea (approval number 2018-04-015). All submitted clinical specimens were decontaminated with the same volume of 2% N-acetyl-l-cysteine-sodium hydroxide and then centrifuged at 3,000 ×
g for 20 minutes. The AFB smear was initially performed with an auramine-rhodamine fluorescent stain, which was confirmed by Ziehl-Neelsen staining. Smear results were graded based on the American Thoracic Society (ATS)/Centers for Disease Control and Prevention guidelines [
1415]. Smear-positive is defined as a grade higher than 1+. Mycobacterial culture was performed by inoculating 500 µL and 100 µL aliquots of decontaminated specimens into a mycobacterial growth indicator tube (MGIT 960 system; Becton Dickinson, Sparks, MD, USA) and onto 3% Ogawa agar (Shinyang, Seoul, Korea) as a reference method. The inoculated media were incubated at 36℃ for six weeks. For culture-positive specimens, we routinely performed GENEDIA MTB/NTM assay for MTBC and NTM differentiation.
To extract DNA, 100 µL of decontaminated sputum specimens were suspended in 100 µL of DNA extraction buffer, and the mixtures were boiled for 10 minutes. After centrifugation, the supernatants were used for both GENEDIA MTB/NTM and GENEDIA MTB assays, which were conducted in parallel, according to the manufacturer's instructions. For the two GENEDIA assays, positive and negative controls were included in each run, and an internal control was used throughout the assay to detect PCR inhibitors. PCR amplification was conducted with a 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). Positive MTBC and NTM results of GENEDIA MTB/NTM were determined when the cycle threshold (Ct) value was less than 40 and 35, respectively. The same Ct value of 40 was used for the MTBC detection using GENEDIA MTB.
The detection limit of the GENEDIA MTB/NTM was measured using serial dilutions of DNA extracted from the reference strains (MTB, Mycobacterium avium, and Mycobacterium abscessus). The lower detection limit was defined as the lowest concentration detected by the assay in a series of three replicates.
Patient information was reviewed in NTM-positive patients to determine the association between the NTM PCR results and NTM-PD. The ATS/Infectious Diseases Society of America diagnostic criteria for NTM disease were applied to patients from whom NTM species were isolated [
16].
The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) with 95% confidence intervals (CIs) of each assay stratified by AFB smear status was calculated based on the results of the reference methods. Kappa value was calculated to compare the agreement between GENEDIA MTB/NTM and GENEDIA MTB for the detection of MTBC. A Chi-square test was used to determine the association between the NTM PCR results and NTM-PD. Statistical analyses were performed using MedCalc Statistical Software version 19.0.5 (MedCalc Software, Ostend, Belgium) and the VassarStats website (
http://vassarstats.net/).
P<0.05 was considered statistically significant.
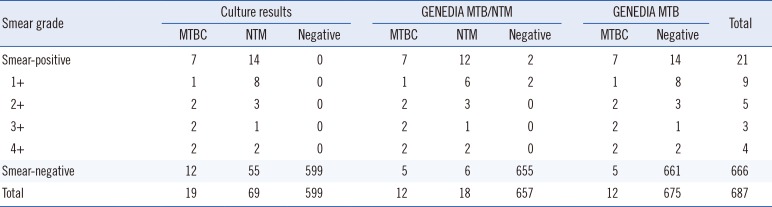
Of the 687 specimens, 12 were positive for MTBC using both the GENEDIA MTB/NTM and GENEDIA MTB assays, with a kappa value of 1.00. Nineteen (2.8%) and 69 (10.0%) specimens were positive for MTBC and NTM by mycobacterial culture, respectively (
Table 1). Seven of the 19 MTBC culture-positive specimens (36.8%) were smear-positive, while the remaining 12 (63.2%) were smear-negative. For the 69 NTM culture-positive specimens, 14 (20.3%) were smear-positive and 55 (79.7%) were smear-negative. Of these culture-positive specimens, 16 were NTM-positive based on the GENEDIA MTB/NTM, while of the 618 NTM culture-negative specimens, two were NTM-positive.
Table 1
GENEDIA MTB/NTM assay, MTB assay, and culture result comparisons stratified by AFB smear grade
|
Smear grade |
Culture results |
GENEDIA MTB/NTM |
GENEDIA MTB |
Total |
|
MTBC |
NTM |
Negative |
MTBC |
NTM |
Negative |
MTBC |
Negative |
|
Smear-positive |
7 |
14 |
0 |
7 |
12 |
2 |
7 |
14 |
21 |
|
1+ |
1 |
8 |
0 |
1 |
6 |
2 |
1 |
8 |
9 |
|
2+ |
2 |
3 |
0 |
2 |
3 |
0 |
2 |
3 |
5 |
|
3+ |
2 |
1 |
0 |
2 |
1 |
0 |
2 |
1 |
3 |
|
4+ |
2 |
2 |
0 |
2 |
2 |
0 |
2 |
2 |
4 |
|
Smear-negative |
12 |
55 |
599 |
5 |
6 |
655 |
5 |
661 |
666 |
|
Total |
19 |
69 |
599 |
12 |
18 |
657 |
12 |
675 |
687 |

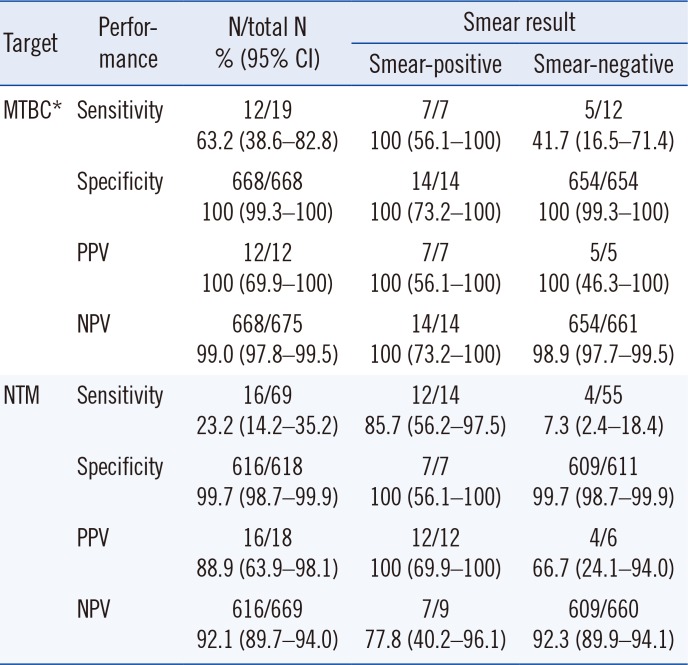
The sensitivities and specificities of the GENEDIA MTB/NTM and GENEDIA MTB assays based on the concurrent culture results with smear status are shown in
Table 2. GENEDIA MTB/NTM and GENEDIA MTB exhibited the same overall performance for MTBC detection from direct sputum specimens. For NTM detection using GENEDIA MTB/NTM, the sensitivity was 85.7% for smear-positive specimens and 7.3% for smear-negative specimens. The PPVs and NPVs for detecting MTBC and NTM are further described in
Table 2, since the population in this study reflects the true disease prevalence in our hospital. Of the 69 NTM culture-positive specimens, 16 were NTM-positive by GENEDIA MTB/NTM, and 62.5% (10/16) were specimens obtained from patients with NTM-PD. The remaining 53 specimens were NTM-negative by GENEDIA MTB/NTM, and 49.1% (26/53) specimens were obtained from patients with NTM-PD. However, the association between NTM PCR-positive results and NTM-PD was not statistically significant (
P=0.35).
Table 2
Diagnostic performance of the GENEDIA MTB/NTM Detection Kit for detecting MTBC and NTM
|
Target |
Performance |
N/total N % (95% CI) |
Smear result |
|
Smear-positive |
Smear-negative |
|
MTBC*
|
Sensitivity |
12/19 63.2 (38.6–82.8) |
7/7 100 (56.1–100) |
5/12 41.7 (16.5–71.4) |
|
Specificity |
668/668 100 (99.3–100) |
14/14 100 (73.2–100) |
654/654 100 (99.3–100) |
|
PPV |
12/12 100 (69.9–100) |
7/7 100 (56.1–100) |
5/5 100 (46.3–100) |
|
NPV |
668/675 99.0 (97.8–99.5) |
14/14 100 (73.2–100) |
654/661 98.9 (97.7–99.5) |
|
NTM |
Sensitivity |
16/69 23.2 (14.2–35.2) |
12/14 85.7 (56.2–97.5) |
4/55 7.3 (2.4–18.4) |
|
Specificity |
616/618 99.7 (98.7–99.9) |
7/7 100 (56.1–100) |
609/611 99.7 (98.7–99.9) |
|
PPV |
16/18 88.9 (63.9–98.1) |
12/12 100 (69.9–100) |
4/6 66.7 (24.1–94.0) |
|
NPV |
616/669 92.1 (89.7–94.0) |
7/9 77.8 (40.2–96.1) |
609/660 92.3 (89.9–94.1) |

The detection limit of GENEDIA MTB/NTM for MTB detection was 5.0×10−6 ng/µL (equal to 1 copy/µL). The measured limits for M. avium and M. abscessus were 5.0×10−3 ng/µL (equal to 953 copies/µL) and 1.5×10−4 ng/µL (27 copies/µL), respectively.
The overall MTBC sensitivity using GENEDIA MTB/NTM was identical to that of the GENEDIA MTB, and all the smear-positive MTBC were detected by both GENEDIA MTB/NTM and GENEDIA MTB. However, smear-negative MTBC specimens showed lower sensitivities with both methods (41.7%), as reported previously in various real-time PCR assays [
1317]. While GENEDIA MTB/NTM detected the vast majority of NTM in smear-positive specimens, NTM sensitivity with smear-negative specimens was <10%. Because differentiation between MTB and NTM is important for smear-negative specimens when mycobacterial infection is suspected, insufficient detection of NTM is a limiting factor for clinical use.
The analytical sensitivity of
M. avium and
M. abscessus was 953 and 27 copies/µL, respectively, with GENEDIA MTB/NTM, suggesting that the clinical sensitivity of NTM could also depend on the species. Further investigation with more focus on the species-specific performance of NTM detection is therefore recommended. In addition, we previously assessed the analytical specificity of GENEDIA MTB/NTM with 85 reference mycobacterial and nonmycobacterial
Corynebacterineae species strains, including one
Corynebacterium sp., two
Gordonia spp., two
Rhodococcus spp., three
Nocardia spp., and one
Tsukamurella sp. [
8]; the assay correctly identified all strains except
Tsukamurella pulmonis.
Recently, Kim, et al. [
18] suggested that an NTM directly isolated from respiratory specimens by real-time PCR assay is a causative organism of true NTM disease. In their study, the vast majority of PCR-positive NTM specimens were obtained from patients with NTM disease. However, our data revealed no significant difference in NTM-PD between the PCR-positive and PCR-negative groups. Further studies are required to confirm the validity of these findings.
This study had some limitations. First, few MTBC culture-positive specimens were included owing to the relatively low number of TB patients enrolled. Second, as only sputum specimens were evaluated, further studies with various types of specimens are required to more broadly evaluate the GENEDIA MTB/NTM assay for differentiation between MTBC and NTM.
In conclusion, our data indicate that the GENEDIA MTB/NTM and the GENEDIA MTB assays have similar clinical performance for detecting MTBC from direct sputum specimens. However, the GENEDIA MTB/NTM assay showed significantly lower sensitivity for NTM detection than for MTBC detection, especially for smear-negative specimens.



