Abstract
Active surveillance culture (ASC) can help detect hidden reservoirs, but the routine use of ASC for extended spectrum β-lactamase-producing Enterobacteriaceae is controversial in an endemic situation. We aimed to determine the prevalence and risk factors of extended spectrum β-lactamase-producing Klebsiella pneumoniae (EBSL-Kpn) colonization among intensive care unit (ICU)-admitted patients. Prospective screening of ESBL-Kpn colonization was performed for ICU-admitted patients within 48 hours for two months. A perirectal swab sample was inoculated on MacConkey agar supplemented with 2 µg/mL ceftazidime. ESBL genotype was determined by PCR-sequencing, and clonal relatedness was evaluated by pulsed-field gel electrophoresis (PFGE). The risk factors of ESBL-Kpn colonization were evaluated. The ESBL-Kpn colonization rate among the 281 patients at ICU admission was 6.4% (18/281), and blaCTX-M-15 was detected in all isolates. ESBL producers also showed resistance to fluoroquinolone (38.9%, 7/18). All isolates had the same ESBL genotype (blaCTX-M-15) and a highly clustered PFGE pattern, suggesting cross-transmission without a documented outbreak. In univariate analysis, the risk factor for ESBL-Kpn colonization over the control was the length of hospital stay (odds ratio=1.062; P=0.019). Routine use of ASC could help control endemic ESBL–Kpn for ICU patients.
Active surveillance culture (ASC) can elucidate hidden reservoirs, which may be delayed or entirely missed in culture results obtained during routine clinical care [1]. To date, there is no consensus regarding the routine use of ASC for extended spectrum β-lactamase-producing Enterobacteriaceae (ESBL-E) in a non-epidemic situation. In most Korean hospitals, ASC has not been routinely recommended because of the low prevalence and cost. However, as the epidemiology changes continuously, the usefulness of ASC should be evaluated considering local epidemiological data and patient vulnerability.
Klebsiella pneumoniae isolates are a common cause of infections in intensive care unit (ICU) patients, and colonizers in the gut could be a source of infection [2]. According to a Korean Global Antimicrobial Resistance Surveillance System (Kor-GLASS) report based on 2017 data, the resistance rate to cefotaxime was 27% in K. pneumoniae blood isolates from May 2016 to April 2017 in six sentinel hospitals [3]. The antimicrobial resistance rate to cefotaxime in K. pneumoniae isolates from ICU patients is higher than that of non-ICU patients [2].
Society for Health Care Epidemiology of America recommends screening methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci carriers, based on early research conducted in the mid-2000s [4]; however, there was not enough evidence for the usefulness of screening ESBL-E [5]. To fill this gap and add to the existing knowledge, we aimed to evaluate the prevalence and risk factors for ESBL-producing K. pneumoniae (ESBL-Kpn) colonization in ICU patients. We hope this data will provide the background evidence for applying routine active surveillance for ESBL-Kpn colonization in high-risk patients as an infection-control policy. The study was approved by the Institutional Review Board of National Health Insurance Service Ilsan Hospital, Goyang, Korea (NHIMC 2018-04-001), which waived the need for informed consent.
From May to July 2016, ESBL-Kpn colonization was screened prospectively within 48 hours for all 281 patients admitted to a surgical or medical ICU at a general hospital in Gyeonggi-do province, Korea. ASC was carried out on perirectal swab samples, which were then inoculated on MacConkey agar supplemented with 2 µg/mL ceftazidime. Identification and susceptibility testing were performed using a MicroScan WalkAway plus system (Beckman Coulter Inc., West Sacramento, CA, USA) and a MicroScan Neg Breakpoint Combo Type 44 panel (Siemens Healthcare Diagnostics Inc., West Sacramento, CA, USA). Antibiotic susceptibility was interpreted using the CLSI guidelines [6]. ESBL-genes were identified by PCR and direct sequencing using gene-specific primers [7]. Cases with a discrepant phenotype and genotype, which showed a positive ESBL result by MicroScan without detection of ESBL genes, were confirmed using a double-disk synergy test [8]. Pulsed-field gel electrophoresis (PFGE) was performed, and the patterns were analyzed using InfoQuest FP software (Bio-Rad, Hercules, CA, USA) to generate a dendrogram based on the unweighted pair-group method, with an arithmetic average (UPGMA) based on Dice's coefficient with 1% band-position tolerance and 0.5% optimization settings [9].
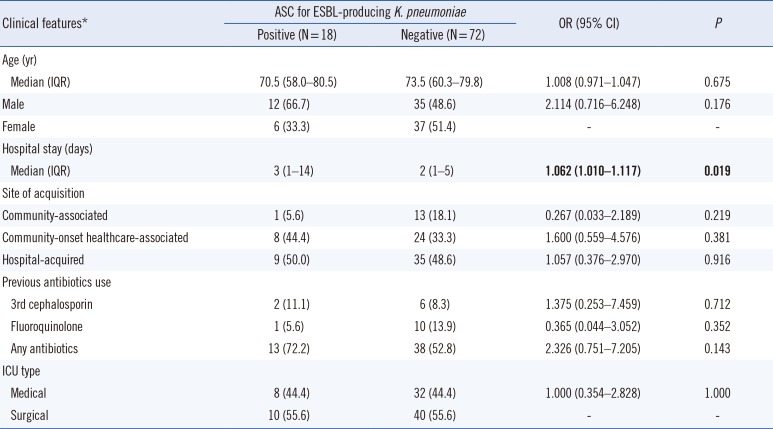
For risk factor analysis, a case-control study with a 1:4 ratio (18 cases and 72 controls) was conducted. The control group included randomly selected patients without ESBL-E isolation in ASC. Clinical information (age, sex, hospital stay, site of acquisition, previous antibiotic use within 90 days before ASC, and ICU types) was collected by reviewing the medical records. Sites of acquisition were identified as described by Friedman, et al. [10] with some modifications. Briefly, hospital-acquired infection/colonization was defined by a positive culture obtained from patients who had been hospitalized for 48 hours or longer. Community-onset infection/colonization was defined by a positive culture obtained at the time of hospital admission or within 48 hours post admission. Community-onset consisted of two groups: community-onset healthcare-associated and community-associated. Community-onset healthcare-associated cases had any of the following histories: attended a hospital or hemodialysis clinic or received intravenous chemotherapy in the last 30 days; hospitalized in an acute-care hospital for two or more days in the last 90 days, or were transferred-in from another healthcare facility before K. pneumoniae isolation. The remaining cases were defined as community-associated.
The age and hospital days were summarized as median, and other variables as N (%) of patients. Continuous variables, such as age, were analyzed using the Mann-Whitney U test. Categorical variables were compared using the chi-squared test to identify independent risk factors. Odds ratio (OR) and 95% confidence interval (CI) were calculated for binomial variables. Variables with P<0.1 in univariate analyses were included, if multivariate logistic regression analysis was needed to identify independent risk factors for ESBL-Kpn colonization. Statistical significance was defined as P<0.05. Data were analyzed using SPSS version 23 (IBM Corp., Armonk, NY, USA).
The ESBL-Kpn colonization rate among the 281 patients at ICU admission was 6.4% (18/281), and blaCTX-M-15 was detected in all isolates. ESBL producers also showed resistance to fluoroquinolone (38.9%, 7/18). The PFGE profiles showed one clustered group and other heterogeneous patterns with 80% similarity (Fig. 1).
In univariate analysis, the only significant risk factor for ESBL-Kpn colonization over the control was hospital stay (OR=1.062; P=0.019). Other factors, such as site of acquisition, previous antibiotic use, and ICU type were not statistically significant (Table 1).
Gastrointestinal colonization by K. pneumoniae has been confirmed as a significant risk factor for infection in ICUs; a half of K. pneumoniae infections originate from patient microbiota [11]. Disruption of normal barriers, for example by gastric acidity and colon microflora, facilitates the overgrowth of pathogens; many mechanisms, including bacterial translocation from the gut and exogenous translocation via contaminated skin, can contribute to these infections [12]. Furthermore, patient with gastrointestinal problems, such as fecal incontinence and diarrhea, could result in the spread of pathogens into the health-care environment, and treatment with antibiotics places a higher selective pressure on colonization by the pathogen [12].
Infections by a multi-drug resistant pathogen, such as ESBL-Kpn, which usually carries resistance to other classes of antibiotics, could fail to respond to treatment, increasing medical costs, prolonging hospital stays, and elevating the socioeconomic burden [13]. K. pneumoniae is one of the most important hospital pathogens, with a high mortality rate, particularly due to multidrug-resistant ESBL-producing strains [14]. Moreover, K. pneumoniae is known to be 3.7-fold more transmissible than Escherichia coli in mathematical modeling analysis, although the underlying factors of this higher transmissibility have not been elucidated [15]. In our study, the prevalence of ESBL-Kpn colonization was 6.4% at ICU admission. In previous studies, ESBL-Kpn carriage rates ranged from 45% (18/40) to 6.9% (24/347) in ICU patients [1617].
The ESBL-Kpn isolates in our study had the same genotype (blaCTX-M-15), and nine of the 14 isolates showed identical PFGE patterns, suggesting cross-transmission without a documented outbreak. A limitation of this study was that we used ceftazidime in ASC, which could result in the low detection of CTX-M-14 type because the hydrolysis of ceftazidime is lower than that of cefotaxime with this CTX-M type. We could not detect carbapenem-resistant K. pneumoniae; although the prevalence of carbapenem-resistant Enterobacteriaceae (CRE) is still low in Korea [18], the recent sharp rise in CRE caused by carbapenemase-producing Enterobacteriaceae (CPE) has created challenges for infection control in Korea [19]. K. pneumoniae is the most common bacterial type of CPE, carrying various types of carbapenemases, such as K. pneumoniae carbapenemase, New Delhi metallo-β-lactamase, and OXA-type carbapenemases [20]. Given that CPE may emerge in Korea, the routine application of ASC not only for ESBL-Kpn but also for CPE for ICU patients should be considered in the near future.
Notes
References
1. Zahar JR, Blot S, Nordmann P, Martischang R, Timsit JF, Harbarth S, et al. Screening for intestinal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in critically ill patients: expected benefits and evidence-based controversies. Clin Infect Dis. 2019; 68:2125–2130. PMID: 30312366.
2. Lee Y, Kim YA, Song W, Lee H, Lee HS, Jang SJ, et al. Recent trends in antimicrobial resistance in intensive care units in Korea. Korean J Nosocomial Infect Control. 2014; 19:29–36.
3. Lee H, Yoon EJ, Kim D, Jeong SH, Won EJ, Shin JH, et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from Kor-GLASS. Euro Surveill. 2018; 23:1800047.
4. Muto CA, Jernigan JA, Ostrowsky BE, Richet HM, Jarvis WR, Boyce JM, et al. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect Control Hosp Epidemiol. 2003; 24:362–386. PMID: 12785411.
5. Thouverez M, Talon D, Bertrand X. Control of Enterobacteriaceae producing extended-spectrum beta-lactamase in intensive care units: rectal screening may not be needed in non-epidemic situations. Infect Control Hosp Epidemiol. 2004; 25:838–841. PMID: 15518025.
6. CLSI. Performance standards for antimicrobial susceptibility testing. 26th ed. CLSI M100S. . Wayne, PA: Clinical and Laboratory Standards Institute;2016.
7. Ryoo NH, Kim EC, Hong SG, Park YJ, Lee K, Bae IK, et al. Dissemination of SHV-12 and CTX-M-type extended-spectrum β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J Antimicrob Chemother. 2005; 56:698–702. PMID: 16141280.
8. Lee K, Cho SR, Lee CS, Chong Y, Kwon OH. Prevalence of extended broad-spectrum beta-lactamase in Escherichia coli and Klebsiella pneumonia. Korean J Infect Dis. 1994; 26:341–348.
9. Park YS, Bae IK, Kim J, Jeong SH, Hwang SS, Seo YH, et al. Risk factors and molecular epidemiology of community-onset extended-spectrum β-lactamase-producing Escherichia coli bacteremia. Yonsei Med J. 2014; 55:467–475. PMID: 24532519.
10. Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, et al. Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med. 2002; 137:791–797. PMID: 12435215.
11. Gorrie CL, Mirceta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis. 2017; 65:208–215. PMID: 28369261.
12. Donskey CJ. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis. 2004; 39:219–226. PMID: 15307031.
13. Martín-Loeches I, Diaz E, Vallés J. Risks for multidrug-resistant pathogens in the ICU. Curr Opin Crit Care. 2014; 20:516–524. PMID: 25188366.
14. Starzyk-Łuszcz K, Zielonka TM, Jakubik J, Życińska K. Mortality due to nosocomial infection with Klebsiella pneumoniae ESBL+. Adv Exp Med Biol. 2017; 1022:19–26. PMID: 28456930.
15. Gurieva T, Dautzenberg MJD, Gniadkowski M, Derde LPG, Bonten MJM, Bootsma MCJ. The transmissibility of antibiotic-resistant Enterobacteriaceae in intensive care units. Clin Infect Dis. 2018; 66:489–493. PMID: 29020273.
16. Ko YJ, Moon HW, Hur M, Park CM, Cho SE, Yun YM. Fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae in Korean community and hospital settings. Infection. 2013; 41:9–13. PMID: 22723075.
17. Kim J, Lee JY, Kim SI, Song W, Kim JS, Jung S, et al. Rates of fecal transmission of extended-spectrum β-lactamase-producing and carbapenem-resistant Enterobacteriaceae among patients in intensive care units in Korea. Ann Lab Med. 2014; 34:20–25. PMID: 24422191.
18. Kim D, Ahn JY, Lee CH, Jang SJ, Lee H, Yong D, et al. Increasing resistance to extended-spectrum cephalosporins, fluoroquinolone, and carbapenem in gram-negative bacilli and the emergence of carbapenem non-susceptibility in Klebsiella pneumoniae: analysis of Korean antimicrobial resistance monitoring system (KARMS) data from 2013 to 2015. Ann Lab Med. 2017; 37:231–239. PMID: 28224769.
19. Jeong SH, Lee KM, Lee J, Bae IK, Kim JS, Kim HS, et al. Clonal and horizontal spread of the blaOXA-232 gene among Enterobacteriaceae in a Korean hospital. Diagn Microbiol Infect Dis. 2015; 82:70–72. PMID: 25702524.
20. Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, et al. Carbapenemase-producing organisms: A global scourge. Clin Infect Dis. 2018; 66:1290–1297. PMID: 29165604.
Fig. 1
Dendrogram of XbaI-restricted DNA of colonizing ESBL-producing Klebsiella pneumoniae isolated from ICU-admitted patients (N=14). Four ESBL-producing K. pneumoniae isolates were excluded because of repeated failure of reculture. Pulsed-field gel electrophoresis was performed with size marker, Lambda Ladders (Promega, Fitchburg, WI, USA).
Abbreviations: ESBL, extended-spectrum beta-lactamase; ICU, intensive care unit.
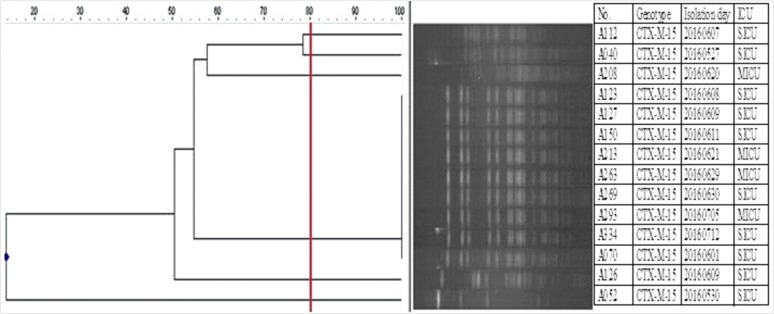
Table 1
Risk factors of ESBL-producing Klebsiella pneumoniae colonization: univariate analysis




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download