Abstract
Background
LINC01234, a long noncoding RNA (lncRNA), is overexpressed in several cancers, including colorectal cancer (CRC). We investigated the role of LINC01234 in CRC development and confirmed its correlation with Krüppel-like factor 6 (KLF6), a tumor suppressor gene that is dysregulated in CRC.
Methods
We tested mRNA levels using quantitative reverse transcription PCR (qRT-PCR). Tissue samples from patients with CRC, inflammatory bowel disease (IBD), hyperplastic polyp, and adenoma were included. Correlations between clinicopathological parameters, overall survival (OS) rate, and LINC01234 were analyzed using Kruskal-Wallis H test. Additionally, cell proliferation, apoptosis, and tumor formation in nude mice were tested to investigate the mechanism of LINC01234. Western blotting was used to determine protein levels.
Results
LINC01234 expression was significantly upregulated in CRC tissues and CRC cell lines than in non-tumor tissues and normal epithelial cells, respectively. LINC01234 was associated with high tumor stage, larger tumor size, and metastasis. Patients with higher LINC01234 expression showed reduced OS. Cell proliferation was inhibited by LINC01234 knockdown, whereas apoptosis was enhanced. Mice injected with SW480 cells with LINC01234 knockdown displayed decreased tumor volume, weight, and Ki-67 levels compared with those injected with control cells. KLF6 was negatively regulated by LINC01234. Overexpression of KLF6 showed effects similar to those observed following LINC01234 knockdown on cell proliferation and apoptosis.
Colorectal cancer (CRC) is the third most malignant tumor in both males and females [1]. CRC incidence and mortality continue to rise in developed countries, possibly due to late diagnosis [2]. Therefore, a sensitive biomarker of CRC progression could dramatically improve poor clinical outcomes.
The encyclopedia of DNA elements (ENCODE) project assessed the entire human genome information and revealed that approximately 80% of the genome performs biochemical functions, whereas only approximately 1.2% is protein-coding [3]. These non-protein-coding genome regions are transcribed into noncoding RNAs, which have wide regulatory functions. Of these RNAs, long noncoding RNAs (lncRNAs), approximately 200 bp to 100 kb in length, mediate numerous cellular activities, such as chromatin remodeling, inhibition of protein interactions, interruption of the splicing process, and modulation of gene transcription by acting as co-activators or repressors [456]. Owing to these varied functions, lncRNAs play important roles in cell growth, differentiation, and homeostasis [78], all of which are known to be disrupted in cancer cells. In fact, dysregulation of lncRNAs has been linked to various cancers [9].
LINC01234 is a recently identified lncRNA that plays important roles in the development of several cancers [10]. High expression of LINC01234 has been observed in numerous tumors, such as gastric cancer [11] and colorectal adenocarcinoma [12]. In triple-negative breast cancer, LINC01234 has been shown to be a core regulatory lncRNA [13]. Furthermore, LINC01234 has been suggested as a potential cancer biomarker owing to its positive correlation with tumor size, tumor stage, and metastasis, as well as a negative correlation with patient survival [1214]. In vitro results have further demonstrated that LINC01234 promotes the proliferation, migration, and invasion and inhibits the apoptosis of esophageal cancer cells [15]. However, little is known regarding the underlying mechanism through which LINC01234 promotes CRC development.
Krüppel-like factor 6 (KLF6) is a transcription factor with a zinc finger structure and is involved in various biological processes, including cell proliferation and differentiation [16]. Loss of KLF6, a tumor suppressor, has been observed in various cancers, including gastric cancer, hepatocellular cancer, lung cancer, and CRC, and it has been demonstrated to play an important role in the early events of cancer development [11171819]. Both KLF6 and LINC01234 have been shown to be critical during the early stage of colon adenocarcinoma (COAD) [20]. Therefore, we investigated whether LINC01234 is associated with CRC and its regulatory effect on KLF6, as well as the functions of KLF6 in CRC.
All CRC cells, including SW480, HCT116, SW620, and LoVo, as well as a normal colonic cell line, NCM460, were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). SW480, SW620, LoVo, and NCM460 were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, New York, NY, USA) containing 10% fetal bovine serum (FBS; Gibco), 100 U/mL penicillin, and 100 µg/mL streptomycin. HCT116 cells were grown in RPMI 1,640 medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. All cells were incubated in a humidified atmosphere at 37℃ and 5% CO2.
The LINC01234 knockdown cell line was established based on a previous report [14]. We transfected cells with scrambled siRNA, si-LINC01234 1#, and si-LINC01234 2# using Lipofectamine 2000 (Invitrogen, Life Technologies, Carlsbad, CA, USA). The medium was changed after four hours of transfection.
Total RNA was isolated using TRIzol (Invitrogen), according to the manufacturer's instructions. For tissue samples, 1 mL of TRIzol reagent was added to 50–100 mg of tissue. For cells, 500 µL of TRIzol reagent were added to 105 cells. Isolation was achieved by adding chloroform to the TRIzol mixture (0.2–1 mL). Next, the colorless upper aqueous phase containing RNA was transferred into a new tube, and the RNA was precipitated by adding 500 µL of isopropanol. The precipitates were washed with 75% ethanol, and the RNA was finally suspended in RNase-free water. cDNA was synthesized using a Reverse Transcription kit (Promega, Madison, WI, USA) with 1 µg of RNA per reaction. qRT-PCR was performed using the SYBR Select Master Mix (Life Technologies), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The comparative cycle threshold (CT; 2−ΔΔCT) method was employed to analyze the results. Each cDNA template had six replicates per experiment, and each experiment was repeated three times. The primers for LINC01234 and KLF6 were as follows: KLF6 forward (F): 5'-CTGCCGTCTCTGGAGGAGT-3', reverse (R): 5'-TCCACAGATCTTCCTGGCTGTC-3'; LINC01234 F: 5'-ACCACACCTGAGATAAGC-3', R: 5'-GAGACAGACAGCAAGAGA-3'; and GAPDH F: 5'-GTCAACGGATTTGGTCTGTATT-3', R: 5'-AGTCTTCTGGGTGGCAGTGAT-3'.
We retrospectively recruited 136 CRC patients who received radical resection at Wuxi People's Hospital Affiliated to Nanjing Medical University, Jiangsu, China, from June 2014 to July 2017. The patients were divided into two groups according to their LINC01234 expression level: high- (N=68) and low-level (N=68) LINC01234. Patient characteristics, including age and gender, were recorded. LINC01234 expression was measured in all 317 colorectal disease tissues obtained from patients diagnosed as having inflammatory bowel disease (IBD; N=49), hyperplastic polyp (N=65), adenoma (N=67), or CRC (N=136) from June 2011 to July 2017. For overall survival (OS) analysis, patients were enrolled from June 2014 and followed up for 60 months, and the OS rate was analyzed using Kaplan-Meier analysis. The study was approved by the Institutional Review Board of Wuxi People's Hospital Affiliated to Nanjing Medical University (Approval No. WPH201104Z), and written informed consent was obtained from all patients. We tested the gene expression of KLF6 and LINC01234 in 76 CRC patients and calculated their correlation.
Tumor location, differentiation, stage, size, and regional lymph node invasion were analyzed and diagnosed using B-mode ultrasound screening. Tissue harmonic and spatial compounding software were employed to reduce noise and increase baseline lesion conspicuity. An ultrasound contrast agent (SonoVue, Bracco, Milan, Italy, sulfur hexafluoride microbubbles) was used to for imaging.
Cell viability was assessed using Cell Counting Kit 8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan), with at least four replicate wells for each group of cells; the results were confirmed by three independent experiments. Cells transfected with si-LINC01234, Scrambled siRNA (si-NC), pCMV-Tag2B-KLF6, or empty vector were seeded in 96-well plates (1,000 or 2,000 cells/well), and CCK-8 values were read every 20 hours. For colony formation, cells were plated on a 6-well plate at a density of 500–1,500 cells/well, with three replicate wells for each group. Methanol was used to fix cells after two weeks of culture, and crystal violet (0.1%) was used for staining; colonies were counted manually. Cell apoptosis was determined by fluorescein isothiocyanate-conjugated annexin V (FITC-annexin V) and propidium iodide (PI) stains and analyzed using FACSCalibur and Cell Quest Pro software (BD Bioscience, San Jose, CA, USA). Briefly, cells were collected using trypsin and washed twice with phosphate-buffered saline (PBS). For staining, 104 cells were suspended in PBS and incubated with 5 µL of FITC-annexin V for 5 minutes and 10 µL of PI for 15 minutes before FACSCalibur analysis.
Protein expression was detected by western blotting, as previously
described [21]. All antibodies, namely those against cleaved caspase 3, poly-ADP-ribose-polymerase (PARP), B-cell lymphoma 2 (Bcl-2), BCL2-associated X (Bax), KLF6, and GAPDH, were purchased from Abcam (Cambridge, MA, USA). The secondary antibody was obtained from Cell Signaling Technology (Danvers, MA, USA). The detection of protein expression was repeated at least three times.
A total of 3×106 SW480 cells transfected with either empty vector or sh-LINC01234 were inoculated subcutaneously to the right flanks of 8-week-old athymic BALB/c nude female mice (weight, ~20 g). A total of 16 mice were used in this study, with eight mice in each group. The animal care and experimental protocols were approved by the Institutional Review Board of Wuxi People's Hospital Affiliated to Nanjing Medical University (Approval No. WPH201104Z). Tumor volume was recorded every five days and calculated as follows: V=0.5×D×d2 (V, volume; D, longitudinal diameter; d, latitudinal diameter). Tumor weight was recorded at day 20 after the mice were sacrificed.
The tumor tissues from mice were stained with Ki-67 (Santa Cruz Biotechnologies, Santa Cruz, CA, USA) using a standard immunohistochemistry protocol, as previously described [22]. All stained sections were analyzed, and two random pictures were chosen for presentation. The protein expression was detected three times using samples collected at different times.
All data are presented as mean±SD. GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA) was used for data analysis. The associations between clinicopathological parameters, OS rate, and LINC01234 expression were analyzed using Kruskal-Wallis H test or Mann-Whitney U test. The Kaplan-Meier method was used to curve the OS rate, followed by the log-rank test for significance comparison. ROC curves (95% confidence intervals [CI]) were employed to evaluate the sensitivity of LINC01234 as a biomarker for distinguishing CRC tissues from benign or normal tissues. The correlation between KLF6 and LINC01234 was calculated using Pearson's correlation analysis. For other comparisons, Student's t-test and one or two-way ANOVAs with the Bonferroni post hoc test were used. P<0.05 was considered statistically significant.
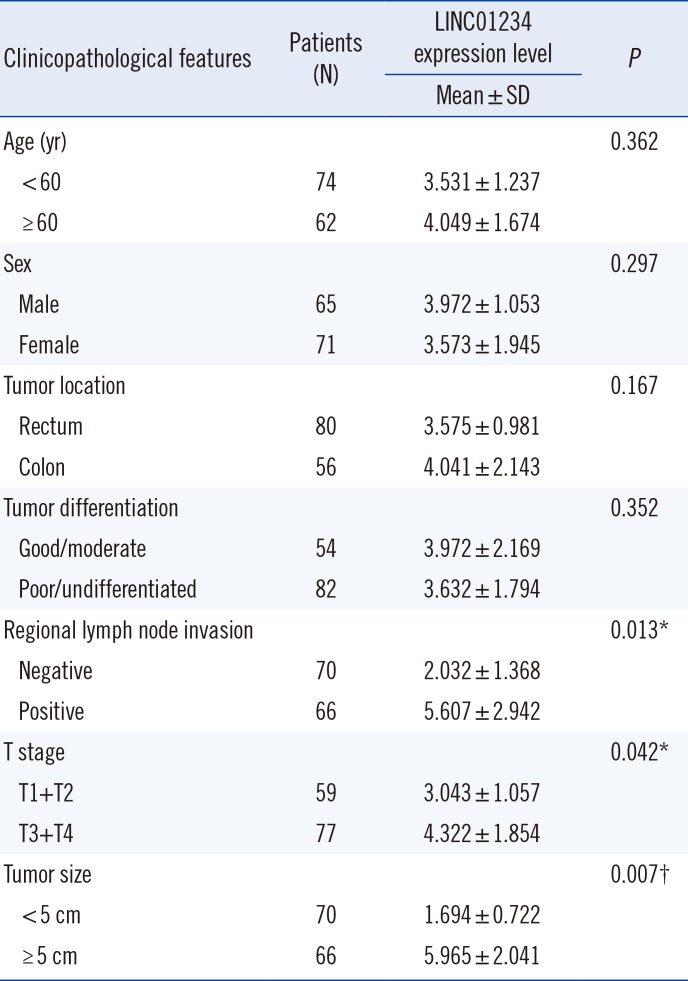
LINC01234 levels were significantly higher in CRC tissue than in other non-tumor tissues (P<0.001; Fig. 1A). However, there was no significant difference across the non-tumor tissues (all P>0.05). To further confirm LINC01234 expression in CRC, we divided the level of LINC01234 in CRC tissues by the level in their paired adjacent non-tumor tissues and calculated the relative expression level using the 2−ΔΔCt method. Most CRC tissues showed high expression of LINC01234 (Fig. 1B). The same LINC01234 trend was observed in CRC cell lines. NCM460 showed a significantly lower level of LINC01234 than other CRC cells, including SW480, HCT116, SW620, and LoVo (Fig. 1C). Furthermore, LINC01234 expression level was associated with pathological characteristics, such as tumor stage (P<0.05), tumor size (P<0.05), and metastasis (P<0.001; Table 1).
We tested the efficiency of LINC01234 knockdown in SW480 and HCT116 cells (Fig. 2A and 2B). LINC01234 knockdown significantly inhibited cell viability and colony formation (Fig. 2C, 2D, and 2E). In contrast, apoptosis rate was higher in LINC01234 knockdown cells than in control cells (Fig. 2F). Correspondingly, several apoptosis-related proteins were upregulated, including cleaved caspase-3, PARP and Bax, whereas Bcl2 was significantly repressed in LINC01234 knockdown cells than in control cells (Fig. 2G).
We confirmed the success of establishment of LINC01234 knockdown cell line (Fig. 3A). Mice injected with LINC01234 knockdown cells showed a significantly smaller tumor volume (Fig. 3B) and lower tumor weight (Fig. 3C) than those injected with control cells. These results suggest that LINC01234 has a significant inhibitory effect on tumor growth, which was further evidenced by the smaller number of Ki-67-positive cells in LINC01234 knockdown cells than in control cells (Fig. 3D).
To further investigate the mechanism of LINC01234 in CRC, we analyzed the correlation between KLF6 and LINC01234 in CRC patients. Intriguingly, KLF6 expression was significantly repressed in CRC tissues than in the adjacent non-tumor tissues; KLF6 was moderately correlated with LINC01234 (R=0.497, P<0.01, Fig. 4A and 4B). KLF6 mRNA and protein levels were significantly increased in LINC01234 knockdown cells than in control cells (Fig. 4C, 4D, and 4E). The same result was also obtained in the LINC01234 knockdown cell line (Fig. 4F).
We established KLF6-overexpressing cell lines, SW480 and HCT116 (Fig. 5A). Overexpression of KLF6 dramatically reduced cell viability and colony formation compared with those of the control cells (Fig. 5B, 5C, and 5D), consistent with the results in the LINC01234 knockdown cells. In addition, apoptosis was enhanced in KLF6-overexpressing cells (Fig. 5E), indicating that LINC01234 plays a role in the regulation of KLF6.
LINC01234 has recently attracted attention because of its high expression levels in different tumors [10], such as triple-negative breast cancer [13], gastric cancer [14], esophageal cancer [15], and colorectal adenocarcinoma [12]. We analyzed a total of 136 patients at different tumor stages and tumor sizes and containing positive or negative regional lymph node invasion. We found that LINC01234 expression level was associated with higher tumor stage and metastasis. These associations have also been observed in gastric cancers [14]. Although LINC01234 is reported to be a biomarker for distinguishing tumor and non-tumor tissues, no correlation between LINC01234 expression and prognosis has been reported [12]. However, LINC01234 was found to predict poorer prognosis in patients with gastric cancer [14]. These different results might be due to the differences in the tumor type and investigation method used. Nevertheless, among a set of regulatory genes, LINC01234 showed a comparatively high k-core score, indicating its key role in breast cancer [13]. Hence, the importance of LINC01234 in tumorigenesis requires further investigation to clarify its mechanism in different tumors.
The association between LINC01234 expression and CRC raised the question of whether LINC01234 impacts cell proliferation or apoptosis. To this end, we knocked down LINC01234 using siRNA; lower LINC01234 expression dramatically decreased cell viability, but increased apoptosis rate and upregulated apoptosis-related proteins. Knockdown of LINC01234 has been reported to induce cell cycle arrest and promote apoptosis in gastric cancer cells [14] and esophageal cancer cells [15]. Furthermore, the levels of several apoptosis-related proteins, including caspase-3, PARP, BAX, and BCL2, were altered in LINC01234 knockdown SW480 and HCT116 cells; similar results have also been reported in gastric cancer cells [14]. To the best of our knowledge, this is the first study showing the effects of LINC01234 in CRC cells. The effects of LINC01234 on cells and tumor formation in nude mice further confirmed that LINC01234 is critical in tumorigenesis. However, further studies including a larger set of CRC cell lines are required to validate these results.
A study examining the association between differentially expressed (DE) microRNA-DElncRNA-DEmRNA interactions and early-stage COAD identified KLF6 and LINC01234 as two important factors related to COAD [20]. To obtain deeper insights into the mechanism of LINC01234, we compared the expression levels of KLF6 in CRC and non-tumor tissues. KLF6 expression was lower in CRC than in non-tumor tissues and it moderately correlated with LINC01234 levels (R=0.497, P<0.01). KLF6 is a known tumor suppressor [232425], and its aberrant expression has been observed in hepatic and gastric cancers [17]. Splice variants of KLF6 have been linked with poor prognosis and are considered risk factors in several cancers such as breast, lung, and prostate cancers [182627].
Early studies demonstrated that KLF6 is frequently inactivated in CRC [28]. A recent study revealed that the mRNA level of KLF-SV2 tended to be reduced in CRC tissues compared with that in adjacent non-tumor tissues [29]. Additionally, overexpression of KLF-SV2 markedly decreased the proliferation and enhanced the apoptotic rate of SW480 and SW620 cells. Those results are consistent with the present results, as well as reports that KLF6 mutants play important roles in CRC development [1630]. KLF6 likely disrupts the complex formation between cyclin D1 and cyclin-dependent kinase 4 (Cdk4), which is critical for cell cycle progression, causing cell cycle arrest [31]. However, KLF-SV2 mutations antagonize the functions of KLF6, thus promoting cell growth. Studies found that KLF6 expression contributes only to tumor growth, and not to metastasis or progression through stages [16], suggesting that dysregulation of KLF6 expression could be an early event in CRC development. It is possible that overexpression of LINC01234 represses KLF6 at the beginning of tumorigenesis, causing uncontrolled cell cycle progression.
Further studies are needed to comprehensively establish the mechanism through which LINC01234 functions in CRC development and modulates KLF6 protein level (e.g., via alteration of KLF6 promoter methylation). However, it is possible that KLF6 is not the key factor promoting CRC development. Therefore, RNA-seq analysis might provide more comprehensive insights into the changes in the gene expression profile.
This study has several limitations. First, we investigated only one specific lncRNA in CRC patients. Other lncRNAs might also be dysregulated in CRC. Second, we did not correlate the LINC01234 expression in each patient with the pathological pattern.
In conclusion, LINC01234 expression was elevated in CRC than in the paired adjacent non-tumor tissues and non-tumor colorectal tissues. This observation was confirmed by comparing LINC01234 levels in CRC cells and normal epithelial cells. LINC01234 was significantly associated with tumor size, stage, and metastasis. Our study provides further support for the use of LINC01234 as a biomarker for predicting poor prognosis of CRC patients. Additionally, the functions of LINC01234 in tumorigenesis appear to mostly occur through the negative regulation of KLF6. Overexpression of KLF6 showed the same trend as LINC01234 knockdown on cell proliferation and apoptosis.
Notes
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018; 68:7–30. PMID: 29313949.
2. Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomarkers Prev. 2016; 25:16–27. PMID: 26667886.
3. Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. PMID: 29126249.
4. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010; 42:1113–1117. PMID: 21057500.
5. Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One. 2012; 7:e47998. PMID: 23133536.
6. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076. PMID: 20393566.
7. Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012; 18:111–123. PMID: 22128341.
8. Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013; 110:2876–2881. PMID: 23382218.
9. Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014; 30:439–452. PMID: 25218058.
10. Cabanski CR, White NM, Dang HX, Silva-Fisher JM, Rauck CE, Cicka D, et al. Pan-cancer transcriptome analysis reveals long noncoding RNAs with conserved function. RNA Biol. 2015; 12:628–642. PMID: 25864709.
11. Ghaleb AM, Yang VW. The pathobiology of Krüppel-like factors in colorectal cancer. Curr Colorectal Cancer Rep. 2008; 4:59–64. PMID: 18504508.
12. He Z, Dang J, Song A, Cui X, Ma Z, Zhang Z. Identification of LINC01234 and MIR210HG as novel prognostic signature for colorectal adenocarcinoma. J Cell Physiol. 2019; 234:6769–6777. PMID: 30362555.
13. Yang F, Liu YH, Dong SY, Yao ZH, Lv L, Ma RM, et al. Co-expression networks revealed potential core lncRNAs in the triple-negative breast cancer. Gene. 2016; 591:471–477. PMID: 27380926.
14. Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018; 24:2002–2014. PMID: 29386218.
15. Ghaffar M, Khodahemmati S, Li J, Shahzad M, Wang M, Wang Y, et al. Long non-coding RNA LINC01234 regulates proliferation, invasion and apoptosis in esophageal cancer cells. J Cancer. 2018; 9:4242–4249. PMID: 30519325.
16. Cho YG, Choi BJ, Song JW, Kim SY, Nam SW, Lee SH, et al. Aberrant expression of Krüppel-like factor 6 protein in colorectal cancers. World J Gastroenterol. 2006; 12:2250–2253. PMID: 16610031.
17. Zhang Q, Tan XP, Yuan YS, Hu CM, He CH, Wang WZ, et al. Decreased expression of KLF6 and its significance in gastric carcinoma. Med Oncol. 2010; 27:1295–1302. PMID: 19967571.
18. DiFeo A, Feld L, Rodriguez E, Wang C, Beer DG, Martignetti JA, et al. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 2008; 68:965–970. PMID: 18250346.
19. Mukai S, Hiyama T, Tanaka S, Yoshihara M, Arihiro K, Chayama K. Involvement of Krüppel-like factor 6 (KLF6) mutation in the development of nonpolypoid colorectal carcinoma. World J Gastroenterol. 2007; 13:3932–3938. PMID: 17663506.
20. Liu JX, Li W, Li JT, Liu F, Zhou L. Screening key long non-coding RNAs in early-stage colon adenocarcinoma by RNA-sequencing. Epigenomics. 2018; 10:1215–1228. PMID: 30182733.
21. Xu Y, Jiang X, Cui Y. Upregulated long noncoding RNA PANDAR predicts an unfavorable prognosis and promotes tumorigenesis in cholangiocarcinoma. Onco Targets Ther. 2017; 10:2873–2883. PMID: 28652769.
22. Prigge ES, Toth C, Dyckhoff G, Wagner S, Müller F, Wittekindt C, et al. p16(INK4a) /Ki-67 co-expression specifically identifies transformed cells in the head and neck region. Int J Cancer. 2015; 136:1589–1599. PMID: 25104331.
23. Cho YG, Kim CJ, Park CH, Yang YM, Kim SY, Nam SW, et al. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005; 24:4588–4590. PMID: 15824733.
24. Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001; 294:2563–2566. PMID: 11752579.
25. DiFeo A, Narla G, Hirshfeld J, Camacho-Vanegas O, Narla J, Rose SL, et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006; 12:3730–3739. PMID: 16778100.
26. Hatami R, Sieuwerts AM, Izadmehr S, Yao Z, Qiao RF, Papa L, et al. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci Transl Med. 2013; 5:169ra12.
27. Narla G, DiFeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005; 65:1213–1222. PMID: 15735005.
28. Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004; 126:1090–1103. PMID: 15057748.
29. Zhang B, Guo DD, Zheng JY, Wu YA. Expression of KLF6-SV2 in colorectal cancer and its impact on proliferation and apoptosis. Eur J Cancer Prev. 2018; 27:20–26. PMID: 29084019.
30. Cho YG, Choi BJ, Kim CJ, Song JW, Kim SY, Nam SW, et al. Genetic alterations of the KLF6 gene in colorectal cancers. APMIS. 2006; 114:458–464. PMID: 16856969.
31. Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, et al. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004; 64:3885–3891. PMID: 15172998.
Fig. 1
LINC01234 is upregulated in CRC tissues and cells and associated with poor prognosis in CRC patients. (A) Quantitative analysis of lncRNA LINC01234. *P<0.001 vs CRC. (B) LINC01234 expression was analyzed in CRC tissues and the corresponding adjacent non-tumor tissues; data are presented as log2 fold change (CRC/NC; N=136). Median value of the log2 fold change is 1.736, and log2 fold changes were defined as follows: >1.736, high expression; <1.736, low expression. (C) Relative expression of LINC01234 in CRC cell lines and normal colon epithelial cell line NCM460 (N=10). †P<0.001 vs NCM460. (D) Patients with high LINC01234 expression (N=68) showed shorter OS (P<0.05) than those with low LINC01234 expression (N=68). (E) Analysis of the sensitivity and specificity of LINC01234 for distinguishing CRC tissues from normal and benign tissues by ROC curve analysis (cut-off=2.601). Data are presented as mean±SD.
Abbreviations: CRC, colorectal cancer; NC, negative control (adjacent non-tumor tissues); IBD, inflammatory bowel disease; HP, hyperplastic polyp; AD, adenoma; OS, overall survival; ROC, receiver operating characteristic; AUC, area under the curve; CI, confidence interval.
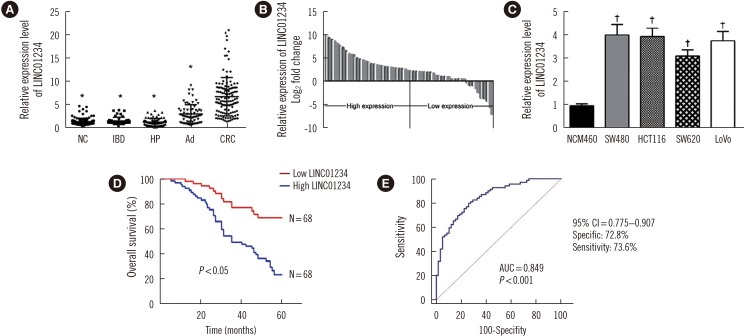
Fig. 2
Effects of LINC01234 knockdown on CRC cell viability and apoptosis in vitro. (A) and (B) LINC01234 expression level was determined by qRT-PCR following transfection of SW480 and HCT116 cells with si-LINC01234 (N=6) or si-NC. *P<0.001 vs Scrambled. (C) and (D) CCK-8 analyses were used to determine the viability of si-LINC01234-transfected SW480 and HCT116 cells. Values are from average of three independent experiments. †P<0.01 vs Scrambled. (E) Colony-forming analyses were conducted to determine the proliferation of si-LINC01234-transfected SW480 and HCT116 cells. Values are from three independent experiments. *P<0.001 vs Scrambled. (F) Flow cytometry was performed to analyze apoptosis rates in CRC cells transfected with si-LINC01234 after 48 hours. Values are from three independent experiments. *P<0.001, †P<0.01 vs Scrambled. (G) Western blot analysis of apoptosis-related proteins after transfection of SW480 and HCT116 cells with Scrambled siRNA, si-LINC01234 1#, or si-LINC01234 2#. GAPDH was used as an internal control. Data are presented as mean±SD.
Abbreviations: CRC, colorectal cancer; qRT-PCR, quantitative reverse transcription PCR; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CCK-8, Cell counting Kit 8.
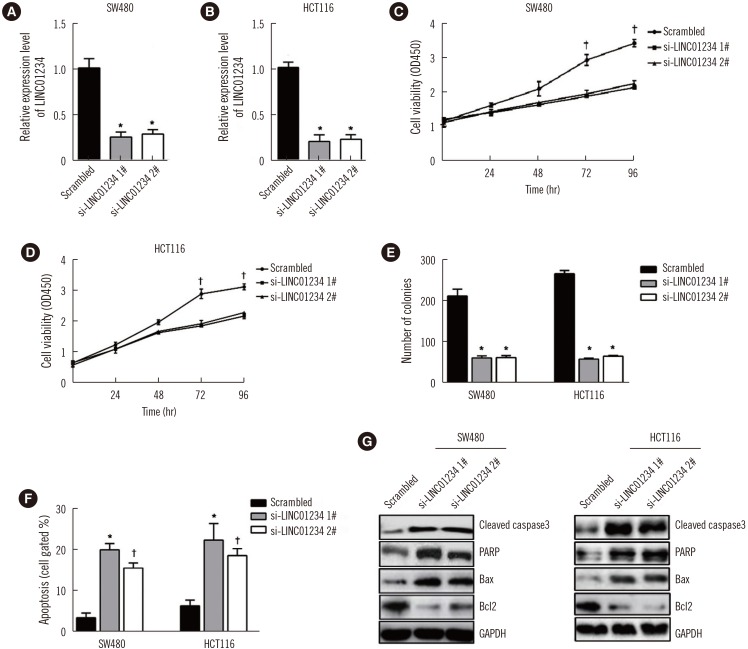
Fig. 3
Down-regulation of LINC01234 suppresses tumor growth in vivo. (A) qRT-PCR analysis of LINC01234 expression in tumor tissues formed from SW480/sh-LINC01234, SW480/empty vector. *P<0.001, sh-LINC01234 vs Empty vector. (B) and (C) Tumor volume was calculated every four days after injection of SW480 cells stably transfected with sh-LINC01234 or empty vector (N=8). *P<0.001, †P<0.01, sh-LINC01234 vs Empty vector. (D) Tumors developed from sh-LINC01234-transfected SW480 cells showed lower Ki-67 protein levels than tumors developed from control cells. Scale bar, 100 µm. Data are presented as mean±SD.
Abbreviation: qRT-PCR, quantitative reverse transcription PCR.
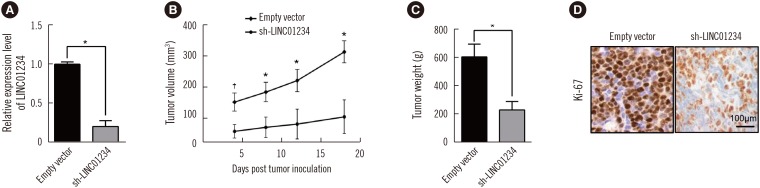
Fig. 4
LINC01234 represses KLF6 expression. (A) KLF6 expression in CRC tissues (N=76) compared with that in corresponding non-tumor tissues (N=76). KLF6 expression was examined by qRT-PCR and normalized to GAPDH expression. *P<0.01, NC vs CRC. (B) Co-expression analysis between LINC01234 and KLF6. (C) KLF6 mRNA levels were detected by qRT-PCR following the transfection of SW480 and HCT116 cells with si-LINC01234; the results were normalized to GAPDH expression. †P<0.001 vs Scrambled. (D) & (E) KLF6 protein levels were determined by western blotting following the transfection of SW480 and HCT116 cells with si-LINC01234. Values are from three independent experiments. †P<0.001 vs Scrambled. (F) KLF6 expression level in mouse tumors was determined by qRT-PCR formed from SW480/sh-LINC01234 or SW480/empty vector (N=8). ‡P<0.001, sh-LINC01234 vs Empty vector. Data are presented as mean±SD.
Abbreviations: KLF6, Krüppel-like factor 6; qRT-PCR, quantitative reverse transcription PCR; CRC, colorectal cancer; NC, negative control (adjacent non-tumor tissues); GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
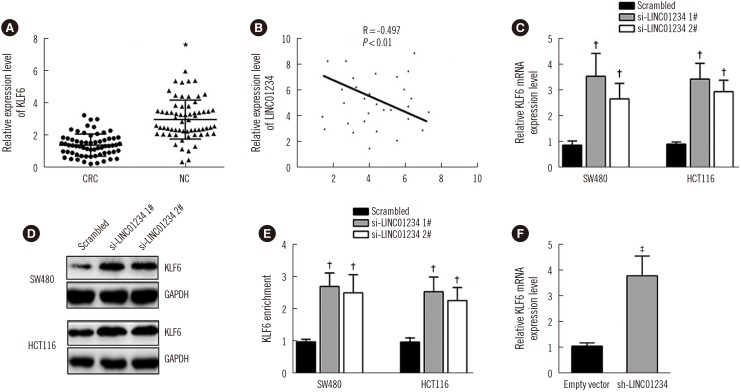
Fig. 5
Overexpression of KLF6 inhibits CRC cell proliferation and increases apoptosis. (A) mRNA level of KLF6 in SW480 and HCT116 cells transfected with pCMV-Tag2B-KLF6 was measured by qRT-PCR. (B)–(D) CCK-8 and colony-forming analyses were used to determine the viability of pCMV-Tag2B-KLF6-transfected SW480 and HCT116 cells. Values are from three independent experiments. (E) Apoptosis was determined by flow cytometry. Values are from three independent experiments. Data are presented as mean±SD. *P<0.001, †P<0.01.
Abbreviations: KLF6, Krüppel-like factor 6; CRC, colorectal cancer; CCK-8, Cell Counting Kit 8; qRT-PCR, quantitative reverse transcription PCR.
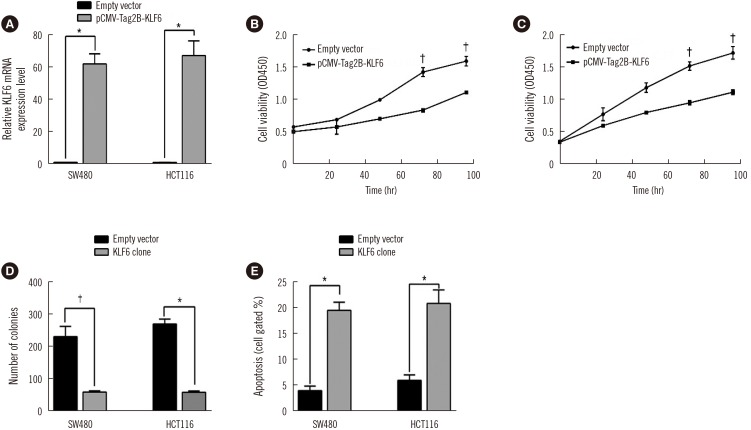
Table 1
Associations between clinicopathological parameters and LINC01234 expression level in patients with CRC (N=136)




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download