Abstract
Mitochondrial dysfunction is closely associated with reactive oxygen species (ROS) generation and oxidative stress in cells. On the other hand, modulation of the cellular antioxidant defense system by changes in the mitochondrial DNA (mtDNA) content is largely unknown. To determine the relationship between the cellular mtDNA content and defense system against oxidative stress, this study examined a set of myoblasts containing a depleted or reverted mtDNA content. A change in the cellular mtDNA content modulated the expression of antioxidant enzymes in myoblasts. In particular, the expression and activity of glutathione peroxidase (GPx) and catalase were inversely correlated with the mtDNA content in myoblasts. The depletion of mtDNA decreased both the reduced glutathione (GSH) and oxidized glutathione (GSSG) slightly, whereas the cellular redox status, as assessed by the GSH/GSSG ratio, was similar to that of the control. Interestingly, the steady-state level of the intracellular ROS, which depends on the reciprocal actions between ROS generation and detoxification, was reduced significantly and the lethality induced by H2O2 was alleviated by mtDNA depletion in myoblasts. Therefore, these results suggest that the ROS homeostasis and antioxidant enzymes are modulated by the cellular mtDNA content and that the increased expression and activity of GPx and catalase through the depletion of mtDNA are closely associated with an alleviation of the oxidative stress in myoblasts.
Mitochondria are an essential organelle not only for maintaining cellular homeostasis and metabolism but also for cell survival and death [1]. Because the mitochondria are the major sites for oxygen reduction, they are confronted with the generation of reactive oxygen species (ROS) from a range of cellular regulation processes and oxidative phosphorylation. In this respect, defects in mitochondrial homeostasis increase superoxide leakage from the electron transport chain (ETC), thereby elevating mitochondrial ROS production, which could lead to oxidative stress and apoptosis [123]. For example, various chemical inhibitors of ETC, such as rotenone and antimycin A, provoke mitochondrial dysfunction, which causes a drastic increase in ROS production [23]. Similarly, the genetic deletion of COX-4 in ETC induces mitochondrial dysfunction and augments the production of ROS [4]. In addition, the cellular treatment of oligomycin, an ATP synthase inhibitor, also caused a drastic increase in the ROS level by inhibiting ETC [5]. Therefore, emerging evidence suggests that mitochondrial dysfunction induced by the inhibition of mitochondrial ETC or ATP synthase can lead to the accumulation of ROS in various cells.
Interestingly, mitochondria possess their own circular DNA, mitochondrial DNA (mtDNA), which contain the genes essential for the crucial proteins for electron transport, oxidative phosphorylation, and mitochondrial homeostasis [6]. For this reason, abnormalities in the quantity and quality of mtDNA are closely associated with mitochondrial dysfunction, oxidative stress, and apoptosis [78]. Studies on the chemical inhibition of mtDNA showed that prolonged exposure to low concentrations (0.1–2 µg/ml) of ethidium bromide (EtBr), an inhibitor of DNA/RNA synthesis, impaired mtDNA replication and transcription without affecting nuclear DNA replication and cell proliferation [910]. Moreover, the mtDNA of myocytes was depleted by prolonged exposure to EtBr, which was reversed by the removal of EtBr [1112]. Therefore, the use of EtBr as a cellular mtDNA depletion/repletion strategy allows a determination of the cellular responses, such as ROS accumulation and antioxidant enzyme change, which are affected by a change in the mtDNA content.
The depletion of mtDNA may increase the superoxide leakage from ETC, which could lead to lethal injury. Several studies have suggested that the complete loss of mtDNA, so-called ρ0 phenotype, by EtBr increased ROS production in a range of cells due to mitochondrial dysfunction and impairment of ETC [1314]. On the other hand, some studies reported that the depletion of mtDNA reduced the level of cellular ROS, which imparted remarkable resistance to oxidative stress and cell death in many cell lines [31516]. Nevertheless, most studies examining the effects of EtBr on ROS production and oxidative stress were analyzed in the ρ0 cell state, which cannot generate ROS and ATP from the mitochondria due to a lack of functional ETC. Therefore, how the ROS level is modulated by the cellular mtDNA content is still a controversial issue. Furthermore, the intracellular ROS level is also maintained by a balance between its production and elimination [17]. The augmentation of ROS and oxidative stress could be restored to physiological levels by regulating the enzymatic and nonenzymatic antioxidant components, such as glutathione peroxidase (GPx), glutathione reductase (GR), catalase, superoxide dismutase (SOD), and reduced glutathione (GSH) [117]. Mitochondrial dysfunction is closely associated with the induction of diverse enzymes involved in the adaptive cellular response via mitochondria-to-nucleus retrograde signaling [181920]. Although an adaptive increase in the expression of antioxidant enzymes allows the cells to resist oxidative stress, it is unclear if or how the altered cellular mtDNA contents modulate the oxidative stress and antioxidant system in myoblasts. Therefore, this study hypothesized that the cellular ROS levels and antioxidant enzymes could correspond to a change in the mtDNA content. The expression and activity of GPx and catalase were found to be inversely correlated with the mtDNA content in myoblasts. A decrease in mtDNA content increased the expression of GPx and catalase and alleviated the ROS accumulation and cell death induced by H2O2, suggesting that the ROS homeostasis and oxidative stress are modulated by the cellular mtDNA content in myoblasts.
L6-GLUT4myc cells, a L6 skeletal muscle cell line (Kerafast, Inc., Boston, MA, USA), were harvested in α-MEM supplemented with a 10% FBS and 1% penicillin-streptomycin solution according to a previous study [12]. As prolonged exposure to low concentrations of EtBr inhibits the replication of mtDNA selectively, a L6-GLUT4myc cell line depleted of mtDNA was prepared by incubating the cells with EtBr (0.2 µg/ml) and uridine (50 µg/ml) for 3 weeks. The reversal of mtDNA depletion was achieved within 7 days by the removal of EtBr and uridine. The control cells were cultured for the same time period under normal harvesting conditions.
Total cellular DNA was extracted by using the DNeasy Tissue kit (Qiagen, Valencia, CA, USA), and total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The reverse transcription reaction was performed using M-MLV reverse transcriptase (Promega, Madison, WI, USA). The primers and PCR conditions used in this study are shown in Table 1. The quantitative real-time RT-PCR (qRT-PCR) amplification was carried out using SYBR green master mix in a LightCycler 480 (Roche Applied Science, Indianapolis, IN, USA).
The myoblasts were treated with the active mitochondrial-specific fluorescent dye, MitoTracker (Molecular Probes, Eugene, OR, USA), for 15 min at 37℃ and washed three times with cold PBS. The fluorescent intensity, visualized, and photographed in a fluorescence microscope (Leica, Germany), correlates with the integrity of mitochondrial function [21].
The intracellular ROS levels were analyzed using 2′,7′-dichlorofluorescin diacetate (DCFH-DA), a cell-permeable oxidationsensitive fluorescent probe, based on the oxidation of DCFH-DA to fluorescent dichlorofluorescein (DCF). Briefly, the cells were treated with 10 µM of DCFH-DA at 37℃ for 30 min and observed by fluorescence microscopy (Carl Zeiss, Göttingen, Germany) because the fluorescence intensity corresponds to the amount of ROS generated in the cells. The fluorescence was determined immediately with excitation and emission wavelengths set at 485 and 530 nm, respectively. The level of mitochondrial ROS was also determined by MitoSOX Red (Life Technology, Carlsbad, CA, USA) dye. Briefly, myoblasts were loaded with 5 µM MitoSOX Red at 37℃ for 30 min. The fluorescence intensity was determined with excitation and emission wavelengths set at 510 nM and 580 nM, respectively. To determine the cellular ATP contents, the cells were pelleted in an ice-cold lysis buffer, and the supernatants were measured by using an ATP Bioluminescence Detection Kit (Promega).
The myoblasts were homogenized with a lysate buffer containing 0.1% Triton X-100 and centrifuged to remove the cell debris and insoluble materials. The activities of the various antioxidant enzymes in the cell lysates were analyzed using a commercially available enzyme assay kit according to the manufacturer's instructions. The GPx, Catalase, GR, and MnSOD enzyme activities were determined quantitatively using enzyme assay kits from BioVision (Mountain View, CA, USA). The kits for measuring the glutathione S-transferase (GST) and glucose-6 phosphate dehydrogenase (G6PD) activities were obtained from Sigma-Aldrich. The GR activity was analyzed using Bioxytech GR-340 from OxisResearch (Portland, OR, USA).
GSH level was measured by using a Glutathione Assay Kit (BioVision). GSH generates a yellow colored complex, 2-nitro-5-thiobenzoic acid, and the absorbance was measured at 405 nm. GSSG level was measured using an OxiSelect Glutathione Assay Kit (Cell Biolabs, Inc., San Diego, CA, USA) according to the manufacturer's protocol.
To analyze the H2O2-induced cytotoxicity in myoblasts, the L6-GLUT4myc cells were incubated with the designated concentration (0–500 µM) of H2O2 for 24 h. The cell viability based on the metabolic activity was determined using an MTT Cell Proliferation Assay Kit (BioVision). The release of LDH resulting from a disruption of the cell membrane integrity was determined by assaying the activity of LDH released into the culture medium using a Cytotoxicity Detection Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's protocol.
The myoblasts were lysed in ice-cold buffer containing 10 mM Tris, 5 mM EDTA, and 1% Triton X-100, and the supernatants after centrifugation were solubilized using a Laemmli solution, subjected to SDS-PAGE, and analyzed for the indicated proteins by immunoblotting, as described elsewhere [22]. Protein expression was determined using ECL Western Blotting Detection Reagents (GE Healthcare, Buckinghamshire, UK). The immunoblot intensities were quantified by densitometry using an analytical scanning system (Alpha Innotech, San Leandro, CA, USA).
The antibodies against catalase (ab16731), G6PD (ab993), GR (ab16801), and MnSOD (ab13533) were purchased from Abcam (Cambridge, UK), and the antibody against GPx (sc-22145) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-β-actin was supplied by Sigma. All other unspecified reagents were obtained from Sigma.
As the prolonged incubation of cells with low dose EtBr is a useful model for examining the cellular processes affected by the mtDNA content, the myoblasts containing varying quantities of mtDNA were prepared by long-term incubation with a low dose of EtBr (0.2 µg/ml) as described in the Methods. As shown in Fig. 1, COX-I and COX-II, which are mtDNA-encoded genes, were barely detected by PCR of the genomic DNA from the cells cultured with EtBr, whereas COX-IV, a nuclear DNA-encoded gene, was not changed in the control, mtDNA-depleted, and -reverted myoblasts. Under these experimental conditions, the mtDNA content was reduced to < 10% of the control by incubation with EtBr, whereas the removal of EtBr in the medium reversed the change in mtDNA content dramatically to the control level within 7 days (Fig. 1A). This shows that prolonged exposure to low EtBr concentration can modulate the cellular mtDNA content without affecting the replication of nuclear DNA in myoblasts. The levels of COX-I, -II, and -IV transcription were also determined in the control, mtDNA-depleted, and -reverted cells by qRT-PCR (Fig. 1B). The mtDNA-depleted cells exhibited very low levels of COX-I and -II mRNAs, which are mtDNA-encoded genes. In contrast, the reverted cells included similar amounts of COX-I and -II mRNA to those of the control cells. In addition, the mRNA of COX-IV, which is encoded by nuclear DNA, was not changed in the mtDNA-depleted or -reverted myoblasts, suggesting that the transcription levels of the genes encoded by mtDNA are in accordance with the cellular mtDNA content in myoblasts. This study further examined whether the depletion of mtDNA provoked mitochondrial dysfunction, which was assessed by the mitochondrial transmembrane potential (ΔΨm) and ATP content, in the cells. Mitotracker, which stains the mitochondria in membrane potential (ΔΨm)-dependent manner [21], was used to evaluate the integrity of the mitochondrial function. As shown in Fig. 1C, dust-like fluorescent granules scattered in the cytoplasm were observed clearly in the control myoblasts. In contrast, in the mtDNA-depleted cells, the intensity of fluorescent granules was dark and almost indistinct, whereas the reverted cells exhibited similar fluorescence intensity to the control cells. This suggests that the depletion of mtDNA disrupted ΔΨm in the myoblasts. As anticipated from the impairment of ΔΨm by mtDNA depletion, the total cellular ATP level was significantly lower in the depleted cells than in the control cells (Fig. 1D). Following mtDNA repletion, however, the total cellular ATP content was similar to that of the control cells in the reverted cells (Fig. 1D).
As the expression of many genes is affected by the retrograde stress signaling from the mitochondria [1123], the differentially expressed genes were evaluated using Affymetrix GeneChip microarrays. The levels of certain antioxidant enzymes were found to be affected by mtDNA depletion (data not shown). Therefore, this study next verified the effect of the cellular mtDNA content on the expression of antioxidant enzymes, such as GR, GPx, GST, G6PD, catalase, and MnSOD, in myoblasts. As shown in Fig. 2A, the depletion of mtDNA increased the mRNA levels of GPx and catalase drastically compared to the control and mtDNA-reverted cells. On the other hand, the transcription of GR, GST, G6PD, and MnSOD were not changed by the depletion or restoration of the mtDNA content. qRT-PCR analysis confirmed the significant upregulation of GPx (10.3-fold) and catalase (8.4-fold) in the mtDNA-depleted myoblasts compared to the control (Fig. 2B). The increased transcription of GPx and catalase in the mtDNA-depleted cells was restored to the control level in the mtDNA-reverted cells, clearly showing that the transcription of GPx and catalase was inversely correlated with the mtDNA content in myoblasts.
Because mRNA of GPx and catalase were increased significantly in the mtDNA-depleted myoblasts, the protein expression and activity of those enzymes were next examined. As shown in Fig. 3, the protein expression and activity of GPx and catalase were increased significantly by mtDNA depletion, whereas the protein levels of GR, G6PD, GST, and MnSOD were unaffected by the depletion or restoration of mtDNA in myoblasts. Moreover, the drastic increase in GPx and catalase were returned to the control levels in the mtDNA-reverted myoblasts, indicating that the expression and activity of GPx and catalase were negatively correlated with the cellular mtDNA level.
Next, the soluble cellular antioxidants, such as GSH and GSSG, were analyzed in the myoblasts, because the expression of antioxidant enzymes was affected by the mtDNA content. As shown in Fig. 4A and B, mtDNA depletion decreased the cellular GSH and GSSG contents slightly but significantly compared to the control. Upon mtDNA repletion, however, the GSH and GSSG contents returned to near the control levels. Despite the lower levels of GSH and GSSG in the depleted cells, the GSH/GSSG ratio, which is considered a good index of the cellular redox status, were similar to the control (Fig. 4C). The cellular activity of GST, an antioxidant enzyme involved in GSH conjugation with reactive oxygen species, was unaffected by the mtDNA contents in the myoblasts (Fig. 4D). The decrease in the GSH and GSSG levels may be associated with the lower demand for the soluble GSH/GSSG pool due to increased GPx and catalase in the mtDNA-depleted myoblasts.
To further explore the interrelationship between the mtDNA content and oxidative stress response, the steady-state levels of intracellular ROS and mitochondrial ROS (mt ROS) were determined in myoblasts as describes in the Methods. The levels of cellular ROS (Fig. 5A) and mt ROS (Fig. 5B) were altered markedly by changes in the mtDNA content corresponding to the modulation of antioxidant enzymes. In the mtDNA-depleted cells, the contents of cellular ROS and mt ROS were drastically reduced as compared to the control level. Upon mtDNA restoration, both the cellular ROS and mt ROS contents returned to near the control levels, suggesting that the cellular mtDNA contents are positively correlated with the cellular and mitochondrial ROS levels in myoblasts. To determine the effects of mtDNA content changes on exogenous oxidative stress, L6-GLUT4myc myoblasts were treated with various amounts of H2O2 (125–500 µM), and the cell viability was measured by MTT reduction and LDH release. As shown in Fig. 5C, at concentrations ranging from 250 to 500 µM, treatment with H2O2 decreased the cell viability significantly in control. Upon exposure to 500 µM H2O2, more than 80% of the myoblasts detached and died. On the other hand, the depletion of mtDNA decreased the level of H2O2-induced cell death significantly compared to those of the control. In addition, when the mtDNA content was returned to near the control level, as shown in the reverted cells, the lethality induced by H2O2 in the myoblasts was similar to the control level. In addition, the release of LDH into the culture medium disrupted the cell membrane integrity, which is an indicator of cell death (Fig. 5D). Corresponding to the result from the MTT reduction assay, LDH release to the medium in H2O2-treated cells increased significantly in a dose-dependent manner. On the other hand, the depletion of mtDNA decreased the H2O2-induced release of LDH dramatically. Thus, these results indicate that mtDNA depletion in myoblasts mitigates oxidative stress-induced cell death as well as ROS accumulation.
Although mitochondrial dysfunction is closely associated with ROS generation in cells, the correlation between the cellular mtDNA content and oxidative stress response has been a controversial issue for a long time. The significant contributions of this study to existing knowledge are followed: (i) the cellular antioxidant enzymes are modulated by changes in the cellular mtDNA content in myoblasts; (ii) the expression and activity of GPx and catalase are inversely correlated with the mtDNA content; (iii) the depletion of mtDNA mitigates the intracellular ROS content and oxidative stress-induced cell death in myoblasts. Therefore, this study reveals that ROS homeostasis and oxidative stress are modulated by the cellular mtDNA content and suggests that the upregulation of GPx and catalase are linked closely to the alleviation of the oxidative stress response by the depletion of mtDNA in myoblasts.
In the present study, the cellular ROS levels and expression of antioxidant enzymes were correlated with the mtDNA content in myoblasts. Although an adaptive increase in antioxidant enzymes allows various cells to alleviate oxidative stress, thereby conferring a survival advantage, it is still unclear how the antioxidant system and ROS are modulated by the altered cellular mtDNA levels. The low level of ROS observed in the mtDNA-depleted cells can be attributed to the upregulation of the glutathione redox system, such as GR, GSH, and GPx, and catalase, because mitochondria developed an intrinsic antioxidant enzyme system that can be modulated reciprocally by cellular oxidative stress [11923]. This study showed that the upregulation of GPx and catalase expression is most prominent in mtDNA-depleted cells. In most cells, the detoxification of H2O2 can be achieved by GPx and catalase [1724]. GPx is a crucial intracellular antioxidant enzyme that reduces oxidative stress by preventing the detrimental accumulation of H2O2 [24]. In conjunction with GPx, catalase is also well known as a scavenger of H2O2 and protects cells from oxidative damage by ROS [17]. In this regard, the protective and anti-apoptotic response against H2O2 found in mtDNA-depleted myoblasts were attributed mainly to the upregulation of GPx and catalase.
The expression and activity of GPx and catalase were negatively correlated with the cellular mtDNA level, whereas those of GR and GST were unaffected. The reactions catalyzed by these enzymes are GSH-dependent mechanisms. In particular, GPx catalyzes the reduction of ROS and peroxides coupled with the oxidation of GSH to GSSG, whereas GR is crucial in GSH/GSSG recycling through replenishment of the cellular GSH pool from GSSG by utilizing NADPH [1724]. GST is involved mainly in the conjugation of GSH with xenobiotics and aldehydes, but it also possesses GPx-like activity to some extent [25]. This study has shown that the cellular GSH and GSSG contents were reduced slightly but significantly in the mtDNA-depleted myoblasts without altering GSH/GSSG ratio. Therefore, the increased expression and activity of GPx together with catalase without the appropriate activation of GR and GST can lead to decreases in both the level of GSH and GSSG. Nevertheless, it is also possible that a low level of ROS in the mtDNA-depleted myoblasts might result in less demand for the soluble glutathione pool.
The molecular mechanism underlying the negative correlation between the cellular mtDNA contents and the expression of GPx and catalase is unclear. Although the regulation of GPx and catalase expression has been studied extensively in transcriptional and post-transcriptional processes, it depends mainly on the transcriptional mechanisms regulated by the cellular redox status from various changes [2426]. In mammalian cells, the modulation of nuclear gene expression in response to mitochondrial dysfunction has been reported with a number of signaling pathways implicated in this retrograde communication [2728]. Indeed, transcription factors, such as ATF2, CEBP/δ, NFAT, CREB, PGC-1α, and NF-κB, participate in mammalian mitochondrial retrograde signaling [1923]. The upregulation of GPx and catalase may be mediated transcriptionally as a part of the retrograde signaling response to oxidative stress via the interactions between their promoters and transcription factors, such as AP-1, P53, PGC-1α, and NF-κB [242930]. Although more studies on the detailed regulation mechanism by the transcriptional modulators of GPx and catalase in mtDNA depletion are warranted, the current study suggests that retrograde signaling derived from mtDNA depletion and mitochondrial dysfunction could be an important regulatory mechanism in the upregulation of GPx and catalase.
Overall, the expression and activity of GPx and catalase were found to be inversely correlated with the mtDNA content in myoblasts. Furthermore, the depletion of mtDNA in myoblasts revealed a decrease in ROS content and drastic alleviation of cell death induced by H2O2. These results suggest that the ROS homeostasis and antioxidant enzymes were modulated by a change in the cellular mtDNA content and that the increased expression and activity of GPx and catalase are closely associated with an alleviation of the oxidative stress in mtDNA-depleted myoblasts.
Figures and Tables
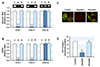 | Fig. 1Alteration of mitochondrial DNA (mtDNA) contents, functional mitochondria, and ATP levels by ethidium bromide (EtBr).(A) The genomic DNA was isolated from the control (C), mtDNA-depleted (D) and -reverted (R) myoblasts, and mtDNA-encoded genes, such as cytochrome c oxidase subunit I (COX-I), subunit II (COX-II), and nuclear DNA-encoded genes, such as cytochrome c oxidase subunit IV (COX-IV), were amplified by PCR. (B) The total RNA was extracted from the myoblasts, and the mRNA levels were quantified by qRT-PCR. The densities were normalized to the β-actin signals, and the relative intensities are expressed in arbitrary units, where the intensity of the control was set to one hundred. ***p < 0.001. (C) To analyze the functional mitochondria, control, mtDNA-depleted and -reverted L6-GLUT4myc myoblasts were stained with MitoTracker. The magnification is ~×400. (D) The total cellular ATP levels were measured by the luciferin-luciferase assay. All results represent the mean ± SEM from five independent experiments. ***p < 0.001 vs. control.
|
 | Fig. 2Effect of mitochondrial DNA (mtDNA) depletion on the transcripts of the antioxidant defense enzymes.The total RNA from the control, mtDNA-depleted (Depleted) and -reverted (Reverted) myoblasts was prepared. The transcript level of the antioxidant defense enzymes was quantified by RT-PCR (A) and qRT-PCR (B). β-Actin was used as the control. All results represent the mean ± SEM from five independent experiments. GR, glutathione reductase; GPx, glutathione peroxidase; GST, glutathione S-transferase; SOD, superoxide dismutase. ***p < 0.001 vs. control.
|
 | Fig. 3Effect of mitochondrial DNA (mtDNA) depletion on the expression and activity of glutathione peroxidase (GPx) and catalase.The total cell lysates were prepared in the control, mtDNA-depleted (Depleted) and -reverted (Reverted) myoblasts. (A–C) The expression levels of GPx and catalase were analyzed by immunoblotting. The densities were normalized to the β-actin signals, and the relative intensities are expressed in arbitrary units, where the intensity of the control was set to one. (D) The total GPx activity was measured using the coupled enzyme procedure with glutathione reductase. The specific activity was calculated using the extinction coefficient obtained from the NADPH standard. (E) The total catalase activity was measured by monitoring the decomposition of 10 mM H2O2 at 240 nm in a medium. One unit of catalase decomposes 1 mM of H2O2 per min. The values are expressed as the mean ± SEM from four independent experiments. GR, glutathione reductase; SOD, superoxide dismutase. ***p < 0.001 vs. control.
|
 | Fig. 4Effect of mitochondrial DNA (mtDNA) depletion on the reduced glutathione (GSH) or oxidized glutathione (GSSG) contents and glutathione S-transferase (GST) activity.(A, B) The cellular GSH and GSSG contents were measured in the control, mtDNA-depleted (Depleted) and -reverted (Reverted) myoblasts. (C) The cellular redox status is presented as the GSH/GSSG ratio. (D) The GST activity was assayed in the supernatant of the cell lysates and was calculated using a known extinction coefficient. The values are expressed as the mean ± SEM from three independent experiments. **p < 0.01 vs. control.
|
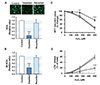 | Fig. 5Effects of mitochondrial DNA (mtDNA) depletion on the cellular reactive oxygen species (ROS) content and H2O2-induced cell death.(A) Control, mtDNA-depleted (Depleted) and -reverted (Reverted) myoblasts were analyzed for the cellular ROS content using a 2′,7′-dichlorofluorescin diacetate (DCFH-DA) probe. The fluorescent images were observed by confocal microscopy (excitation, 485 nm; emission, 530 nm). The relative fluorescence intensities obtained from 50 cells are expressed in arbitrary units where the intensity of the control was set to one. (B) The level of mitochondrial ROS (mt ROS) was also determined by MitoSOX. The fluorescence intensity was determined with excitation (510 nM) and emission (580 nM) wavelengths. (C, D) Control (circle), mtDNA-depleted (triangle) and -reverted (rectangle) myoblasts were incubated with 0, 25, 250, or 500 mM H2O2 for 24 h. The effects of H2O2-induced cell death were evaluated by MTT reduction assay (C) and LDH release (D) as described in the Methods. The results of MTT reduction are expressed as the percentage change where the absorbance of myoblasts without H2O2 was set to one hundred. LDH release was determined by assaying the activity of LDH released into the culture medium. All results represent the mean ± SEM from four independent experiments. ***p < 0.001; **p < 0.01 vs. control.
|
ACKNOWLEDGEMENTS
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (2016R1D1A1B03933506, 2017M2B2A4049415) and Research Grant for Outstanding Researchers by the Dongguk University Research Fund (K-2018-G0002-00016).
Notes
References
1. Schulz E, Wenzel P, Münzel T, Daiber A. Mitochondrial redox signaling: interaction of mitochondrial reactive oxygen species with other sources of oxidative stress. Antioxid Redox Signal. 2014; 20:308–324.

2. Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003; 278:8516–8525.

3. Mercy L, Pauw Ad, Payen L, Tejerina S, Houbion A, Demazy C, Raes M, Renard P, Arnould T. Mitochondrial biogenesis in mtDNA-depleted cells involves a Ca2+-dependent pathway and a reduced mitochondrial protein import. FEBS J. 2005; 272:5031–5055.
4. Leadsham JE, Sanders G, Giannaki S, Bastow EL, Hutton R, Naeimi WR, Breitenbach M, Gourlay CW. Loss of cytochrome c oxidase promotes RAS-dependent ROS production from the ER resident NADPH oxidase, Yno1p, in yeast. Cell Metab. 2013; 18:279–286.

5. Chang CJ, Yin PH, Yang DM, Wang CH, Hung WY, Chi CW, Wei YH, Lee HC. Mitochondrial dysfunction-induced amphiregulin upregulation mediates chemo-resistance and cell migration in HepG2 cells. Cell Mol Life Sci. 2009; 66:1755–1765.

6. Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981; 290:457–465.

8. Duchen MR. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol Aspects Med. 2004; 25:365–451.

9. Zylber E, Vesco C, Penman S. Selective inhibition of the synthesis of mitochondria-associated RNA by ethidium bromide. J Mol Biol. 1969; 44:195–204.
10. Desjardins P, Frost E, Morais R. Ethidium bromide-induced loss of mitochondrial DNA from primary chicken embryo fibroblasts. Mol Cell Biol. 1985; 5:1163–1169.

11. Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of interorganelle crosstalk. EMBO J. 1999; 18:522–533.
12. Park SY, Choi GH, Choi HI, Ryu J, Jung CY, Lee W. Depletion of mitochondrial DNA causes impaired glucose utilization and insulin resistance in L6 GLUT4myc myocytes. J Biol Chem. 2005; 280:9855–9864.

13. Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, Lee MS. Resistance of mitochondrial DNA-depleted cells against cell death: role of mitochondrial superoxide dismutase. J Biol Chem. 2004; 279:7512–7520.
14. Nacarelli T, Azar A, Sell C. Inhibition of mTOR prevents ROS production initiated by ethidium bromide-induced mitochondrial DNA depletion. Front Endocrinol (Lausanne). 2014; 5:122.

15. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011; 12:222–230.

16. Perez MJ, Gonzalez-Sanchez E, Gonzalez-Loyola A, Gonzalez-Buitrago JM, Marin JJ. Mitochondrial genome depletion dysregulates bile acid- and paracetamol-induced expression of the transporters Mdr1, Mrp1 and Mrp4 in liver cells. Br J Pharmacol. 2011; 162:1686–1699.

17. Nickel A, Kohlhaas M, Maack C. Mitochondrial reactive oxygen species production and elimination. J Mol Cell Cardiol. 2014; 73:26–33.

18. Behan A, Doyle S, Farrell M. Adaptive responses to mitochondrial dysfunction in the rho degrees Namalwa cell. Mitochondrion. 2005; 5:173–193.
19. Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004; 14:1–15.
20. Brambilla L, Cairo G, Sestili P, O'Donnel V, Azzi A, Cantoni O. Mitochondrial respiratory chain deficiency leads to overexpression of antioxidant enzymes. FEBS Lett. 1997; 418:247–250.

21. Chen LB. Fluorescent labeling of mitochondria. Methods Cell Biol. 1989; 29:103–123.
22. Ryu HS, Park SY, Ma D, Zhang J, Lee W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS One. 2011; 6:e17343.

23. Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001; 20:1910–1920.

24. Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2011; 15:1957–1997.

25. Halliwell B. Antioxidant defence mechanisms: from the beginning to the end (of the beginning). Free Radic Res. 1999; 31:261–272.

26. St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006; 127:397–408.

28. da Cunha FM, Torelli NQ, Kowaltowski AJ. Mitochondrial retrograde signaling: triggers, pathways, and outcomes. Oxid Med Cell Longev. 2015; 2015:482582.

29. Hussain SP, Amstad P, He P, Robles A, Lupold S, Kaneko I, Ichimiya M, Sengupta S, Mechanic L, Okamura S, Hofseth LJ, Moake M, Nagashima M, Forrester KS, Harris CC. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res. 2004; 64:2350–2356.

30. Zhou LZ, Johnson AP, Rando TA. NF kappa B and AP-1 mediate transcriptional responses to oxidative stress in skeletal muscle cells. Free Radic Biol Med. 2001; 31:1405–1416.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download