Abstract
Retinoblastoma (Rb) is one of the most common eye malignancies occur in childhood. The crucial roles of non-coding RNAs, particularly long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), have been widely reported in Rb progression. In the present study, we found the expression of lncRNA T-cell leukemia/lymphoma 6 (TCL6) was significantly downregulated in Rb tissues and cell lines. Knockdown of lncRNA TCL6 promoted cell proliferation while reduced cell apoptosis in Rb cells. Moreover, lncRNA TCL6 serves as a sponge for miR-21, a previously-reported oncogenic miRNA in Rb, by direct targeting to negatively regulated miR-21 expression, therefore modulating Rb proliferation through miR-21. TCL6 overexpression inhibited Rb cell proliferation while miR-21 overexpression exerted an opposing effect; the effect of TCL6 overexpression was partially attenuated by miR-21 overexpression. PTEN/PI3K/AKT signaling pathway was involved in lncRNA TCL6/miR-21 axis modulating Rb cell proliferation. Taken together, lncRNA TCL6 serves as a tumor suppressor by acting as a sponge for miR-21 to counteract miR-21-mediated PTEN repression.
Retinoblastoma (Rb) is known as one of the important childhood malignancies, which is due to inactivation of the Rb gene (tumor suppressor gene). The early detection of Rb could provide better treatment for Rb patients [1]. Timely diagnosis and successful treatment are vital for the improvement of survival rates in patients with Rb.
As is widely known, the PTEN/PI3K/AKT signaling pathway is a highly conserved developmental pathway [2], which is important in the regulation of cellular proliferation, differentiation, and apoptosis in tumors [3]. In Rb, the PTEN/PI3K/AKT signaling also plays a key role in mediating cell proliferation [45], apoptosis [6] and invasion [5]. Inactivation of PTEN, or constitutive activation of PI3K or AKT, suppresses Rb/E2F driven apoptosis in the retina and contributes to Rb formation in mice [7]. Understanding the mechanism of PTEN/PI3K/AKT signaling dysregulation will help to choose a better and effective therapeutic approach.
Among the various biomarkers, non-coding RNAs, mainly long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), have emerged as attractive tools for detecting Rb in various stages [189]. miR-21, the first detected as an oncomiR [10], is commonly overexpressed in cancer to promote cancer cell proliferation, invasion, and migration, such as in breast cancer [11], lung cancer [12], gastric cancer [13] and Rb [414]. Interestingly, miR-21 exerts its biological role in Rb through the PTEN/PI3K/AKT signaling. miR-21 inhibition in Rb cell lines suppressed cell viability, cell migration, and invasion but improved cell apoptotic rates and significantly affected the expression of PTEN, PI3K, and p-AKT [4]. Regarding the mechanism, the activity of miRNA on downstream targets may be counteracted by lncRNAs [15], which inspired us to investigate whether lncRNAs play a potential role in Rb through miR-21 and PTEN signaling.
In the present study, several online tools were employed to identify lncRNAs that might target miR-21 to regulate its expression and effect on downstream PTEN/PI3K/AKT. Of the selected lncRNAs, lncRNA TCL6 has been reported to be downregulated in several carcinomas [1617] (GSE53757 and GSE46699), thus, the function of lncRNA TCL6 in Rb and its correlation with miR-21 and downstream PTEN/PI3K/AKT signaling was further evaluated. Taken together, we demonstrated a novel mechanism by which Rb cell proliferation can be modulated from the perspective of lncRNA-miRNA interaction.
Twenty-two cases human Rb tissues were obtained from patients with Rb at The First Affiliated Hospital of Human Normal University/Hunan Provincial People's Hospital (Changsha, China) with the approval of Human Normal University (from April 2012 to April 2016, No. 2012015). All cases were histopathologically diagnosed as having Rb without radiotherapy or chemotherapy before a surgical operation. The normal retina tissues were obtained from ruptured globes of 8 patients as controls. The written informed consents were obtained from all the patients. All biopsy specimens were immediately frozen in liquid nitrogen and stored at −80℃ before further use.
Human Rb cell lines, Y79 (ATCC HTB-18) and WERI-Rb-1 (ATCC HTB-169) were obtained from ATCC (Manassas, VA, USA) and were both cultured in RPMI-1640 with 20% FBS at 37℃ with 5% CO2. ARPE-19 (ATCC CRL-2302), a retinal pigmented epithelium cell line, was purchased from ATCC and cultured in DMEM: F12 Medium with 10% FBS at 37℃ with 5% CO2.
Expression of miR-21 was achieved in cell lines was achieved by transfection of miR-21 mimics (Ji Ma Pharmaceutical Co., Ltd., Shanghai, China) with the help of Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) according to the protocols. Knockdown or overexpression of lncRNA TCL6 was achieved by transfection of small interfering RNA (si-)-TCL6 or TCL6 overexpressing vector (GeneCopoecia, Guangzhou, China) using Lipofectamine 2000 (Invitrogen).
Total RNA was extracted using Trizol reagent (Invitrogen) following the protocol. The SYBR green PCR Master Mix (Qiagen, Hilden, Germany) was used for mRNA expression detection following the protocol using GAPDH expression as an endogenous control. The expression of miRNA was examined by a Hairpinit™ miRNAs qPCR kit (Genepharma, Shanghai, China) using RNU6B as an endogenous control. The 2−ΔΔCT method was used to analyze the relative fold changes.
MTT assay was performed to evaluate cell viability. 24 h after seeding into 96-well plates (5 × 103 cells/well), cells were transfected and/or treated as described. Forty-eight hours after transfection, 20 µl MTT (at a concentration of 5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added, and the cells were incubated for an additional 4 h in a humidified incubator. 200 µl DMSO was added after the supernatant discarded to dissolve the formazan. OD490 nm value was measured. The viability of the non-treated cells (control) was defined as 100%, and the viability of cells from all other groups was calculated separately from that of the control group.
DNA synthesis was determined according to the incorporation of the thymidine analogue 5-ethynyl 2-deoxyuridine (EDU) into genomic DNA by using the Click-IT EdU Alexa Fluor 488 kit for flow cytometry (Invitrogen). Cells were treated or transfected, and growth medium was replaced with medium containing EDU (10 µmol/L). Two hours later, the medium was replaced with EDU–free medium. Then an Apollo staining and DAPI staining were performed to detect the EDU positive cells with a fluorescence microscope following the protocols. The EDU incorporation rate calculated as the ratio of EdU-positive to total DAPI-positive cells (blue cells).
Cell apoptosis was evaluated with Annexin V-FITC/PI Apoptosis Detection Kit (Vazyme, Nanjing, China) in accordance with the manufacturer's protocol. The cells were analyzed by using a BD FACSCanto II (BD Biosciences, San Diego, CA, USA) flow cytometry and the results were analyzed by FlowJo software (Tree Star Software, San Carlos, CA, USA).
Human lncRNA TCL6 sequence or the mutant sequence of lncRNA TCL6 with the predicted target sites was cloned into the downstream of the Renilla psiCHECK2 vector (Promega, Madison, WI, USA). HEK293 cells were plated onto 24-well plates (5 × 105 cells/well) the day before transfection and were co-transfected with luciferase reporter vectors (0.12 µg) and 40 nM of miR-21 mimics or negative control (NC mimics) by using Lipofectamine 2000 (Invitrogen). Luciferase report assay was measured in Victor 1420 Multilabel Counter (Wallac, Turku, Finland) by using the Luciferase Assay System (Promega) according to the standard protocol.
The protein levels of PTEN, p-AKT, and AKT were examined by immunoblotting. Target cells in lysed using RIPA buffer with 1% PMSF; the proteins were extracted and analyzed for protein concentration using the bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Haimen, Jiangsu, China). Proteins were then loaded onto an sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE) minigel and transferred onto PVDF membrane. Thereafter, the membrane was probed with the antibodies listed below: anti-PTEN (ab32199, Abcam, Cambridge, MA, USA), anti-p-PTEN (ab131107), anti-AKT (ab32505, Abcam), anti-p-AKT (ab81283, Abcam) and GAPDH (ab8245, Abcam) at 4℃ overnight. Thereafter, the blots were incubated with the HRP-conjugated secondary antibody (1:5,000). Signals visualization was conducted by ECL Substrates (Millipore, MA, USA) using GAPDH as an endogenous protein for normalization. The gray intensity analysis was performed using ImageJ software (National Institutes of Health; http://rsb.info.nih.gov/ij/).
The activity of PI3K was measured using ELISA kits (Promega) according to the manufacturer's instructions. Each experiment was repeated three times.
Data are processed using SPSS Statistics 17.0.1 (SPSS Inc., Chicago, IL, USA) statistical software and presented as the mean ± standard deviation of results from at least three independent experiments. A Student t-test was used for statistical comparison between means where applicable. Differences among more than two groups in the above assays were estimated using one-way ANOVA. A p < 0.05 was considered statistically significant.
Since lncRNA TCL6 has been reported to be low-expressed in renal clear cell carcinoma [1617], here, the expression of lncRNA TCL6 was first detected in Rb tissues and cell lines. As shown in Fig. 1A, lncRNA TCL6 expression was significantly downregulated in 22 cases Rb tissues, compared to that in normal retina tissues. Moreover, lncRNA TCL6 was also downregulated in two Rb cell lines, Y79 and WERI-Rb-1, compared to that in a normal retinal pigmented epithelium cell line, ARPE-19 (Fig. 1B). The downregulation of lncRNA TCL6 suggests its potential role in Rb.
To evaluate the molecular function and mechanism of lncRNA TCL6 on Rb progression, Y79 and WERI-Rb-1 cells were transfected with siTCL6 to achieve lncRNA TCL6 knockdown using NC vector as a negative control, as confirmed by real-time PCR (Fig. 2A). Next, the cell viability, cell apoptosis and DNA synthesis capacity of these two cell lines were detected using MTT, Flow cytometry, and EDU assays. Knockdown of lncRNA TCL6 significantly promoted the cell viability and DNA synthesis capacity while reduced the cell apoptosis of Rb cell lines (Fig. 2B–D), indicating that lncRNA TCL6 knockdown may contribute to Rb progression.
As predicted by online tools, lncRNA TCL6 may bind to miR-21 to regulate its expression, therefore modulating Rb cell behaviors through miR-21 downstream PTEN signaling. Here, we investigated whether lncRNA TCL6 serves as a sponge for miR-21 via direct binding. First, we determined the transfection efficiency. As Fig. 3A and B shown, TCL6 overexpression vector and miR-21 mimics transfection effectively increased the TCL6 mRNA and miR-21 levels in Y79 and WERI-Rb-1 cells, respectively. Contrary to lncRNA TCL6, miR-21 expression was significantly upregulated in two Rb cell lines (Fig. 3C). In these two Rb cell lines, miR-21 expression was negatively regulated by lncRNA TCL6 (Fig. 3D, E).
To validate the predicted binding between lncRNA TCL6 and miR-21, two types of luciferase reporter gene vectors, wild-type (wt) and mutant-type (mut) TCL6, were constructed and named wt-TLC6 and mut-TCL6. These two types of vectors contained wild or mutated miR-21 binding site (Fig. 3D). HEK293 cells were co-transfected with vectors and miR-21 mimics and examined for luciferase activity. As shown in Fig. 3E, the luciferase activity of wt-TCL6 was significantly suppressed by miR-21; after mutating the predicted binding site, the alterations of the luciferase were abolished. These findings indicate that lncRNA TCL6 serves as a sponge for miR-21 to negatively regulate its expression.
After confirming the binding between lncRNA TCL6 and miR-21, the dynamic effect on Rb cell proliferation was examined. Y79 and WERI-Rb-1 cells were co-transfected with lncRNA TCL6 overexpressing vector and miR-21 mimics and then examined for cell viability and DNA synthesis capacity. As shown in Fig. 4A, 48 h or 72 h after transfection, the cell proliferation of Rb cells could be significantly inhibited by lncRNA TCL6 overexpression while promoted by miR-21 overexpression; lncRNA TCL6 overexpression-induced cell proliferation inhibition was partially restored by miR-21 mimics (Fig. 4A). Similarly, the EdU assays confirmed miR-21 mimics could reverse TCL6 overexpression-induced inhibition of DNA synthesis capacity (Fig. 4B). These findings indicate that lncRNA TCL6 modulates Rb cell proliferation through miR-21.
Since miR-21 has been reported to regulate Rb progression through PTEN/PI3K/AKT signaling pathway, here, we investigated the effect of lncRNA TCL6/miR-21 axis on PTEN, PI3K, AKT, and p-AKT. In both Rb cell lines, lncRNA TCL6 overexpression significantly increased PTEN and p-PTEN protein levels while reduced p-AKT protein levels; miR-21 mimics reduced PTEN and p-PTEN protein levels while increased p-AKT protein levels; the effect of lncRNA TCL6 overexpression was partially attenuated by miR-21 mimics (Fig. 5A). Meanwhile, the protein levels of AKT were not significantly altered by lncRNA TCL6 overexpression or miR-21 mimics (Fig. 5A). To validate whether lncRNA TCL6 could counteract miR-21-mediated PTEN repression, the protein level of PTEN and p-PTEN in response to TCL6 knockdown was detected. As shown in Fig. 5B, si-TCL6 obviously reduced PTEN and p-PTEN protein levels while increased p-AKT protein levels in Y79 and WERI-Rb-1 cells. The activity of PI3K was determined by ELISA. As Fig. 5C and D shown TCL6 overexpression inhibited the PI3K activity while miR-21 mimics increased the activity. The activity of PI3K was increased by TCL6 knockdown (Fig. 5D). These results indicated that lncRNA TCL6 counteracts miR-21-mediated PTEN repression through serving as a sponge for miR-21.
To further confirm the above findings, the expression of miR-21 and PTEN was detected in tissue specimens. In Rb tissue specimens, miR-21 expression was significantly upregulated while PTEN mRNA expression was downregulated (Fig. 6A, B). LncRNA TCL6 was negatively correlated with miR-21 and was positively correlated with PTEN (Fig. 6C, D). These findings indicate that rescuing lncRNA TCL6 expression in Rb may upregulate PTEN through miR-21, therefore suppressing hyperproliferation of Rb cells.
In the present study, we demonstrated that lncRNA TCL6 expression was significantly downregulated in Rb tissues and cell lines, and lncRNA TCL6 knockdown promoted cell proliferation while inhibited cell apoptosis of Rb cell lines. LncRNA TCL6 served as a sponge for miR-21 to counteract miR-21-mediated PTEN suppression, therefore modulating Rb cell proliferation through PTEN/PI3K/AKT signaling.
The essential role of lncRNAs in carcinogenesis has been widely reported. Generally, lncRNAs are involved in a wide range of biological processes during carcinogenesis, including cell proliferation, invasion, migration, and apoptosis [1819]. In Rb, lncRNAs also represent potential biomarkers and modulate the development. LncRNA brain-derived neurotrophic factor antisense (BDNF-AS) is downregulated in Rb and low BDNF-AS expression is associated with shorter overall survival and may be acting as an independent marker in Rb. Forced overexpression of BDNF-AS in Rb cell lines can inhibit cell proliferation and migration [20]. Another lncRNA, H19, suppresses Rb progression through counteracting miR-17-92 cluster to inhibit Rb cell proliferation, induces retinoblastoma cell cycle arrest and cell apoptosis [21]. In the present study, we found that lncRNA TCL6, which was downregulated in renal clear cell carcinoma [1617] (GSE53757 and GSE46699) and was predicted to target miR-21, was significantly underexpressed in Rb tissue specimens and cell lines, suggesting that lncRNA TCL6 may play a role similar to BDNF-AS and H19. Further functional experiments confirmed that lncRNA TCL6 knockdown promoted Rb cell proliferation while inhibited cell apoptosis, indicating that lncRNA TCL6 served as a tumor suppressor in Rb.
Regarding the underlying mechanism, lncRNAs commonly exert their biological roles through acting as sponges for miRNAs to counteract miRNA-mediated repression of their downstream targets or signaling pathways [15]. Previously, miR-21 has been reported to possess the oncogenic potential to target tumor suppressor PDCD4 and regulate tumor progression and metastasis in Rb [14]. Moreover, miR-21 inhibitor has been reported to suppress Rb cell viability while promote cell apoptosis through PTEN/PI3K/AKT signaling pathway [4]. Here, we used online tools to identify a possible miR-21 binding site in lncRNA TCL6. In Rb cell lines, miR-21 expression was remarkably upregulated, which was consistent with previous studies [414]. Through direct binding, lncRNA TCL6 negatively regulated miR-21 expression. Moreover, lncRNA TCL6 overexpression suppressed Rb cell proliferation while miR-21 overexpression exerted an opposing effect, and the effect of TCL6 overexpression was partially attenuated by miR-21 overexpression. These findings indicate that lncRNA TCL6 modulates Rb proliferation through targeting miR-21 to suppress its oncogenic effect on Rb cells.
As has been reported by previous studies, PTEN/PI3K/AKT signaling plays a key role in Rb progression. PTEN-induced growth arrest in Rb cells can be rescued by the co-expression of active PI3K and downstream effectors such as AKT [22]. PTEN serves as a direct downstream target of miR-198 to participate in miR-198 modulating Rb cell proliferation [5]. More importantly, miR-21 also exerts its oncogenic effect on Rb progression through PTEN/PI3K/AKT signaling pathway [4]. In the present study, lncRNA TCL6 overexpression obviously increased PTEN protein level while reduced p-AKT protein level. On the contrary, miR-21 overexpression exerted an opposing effect on PTEN and p-AKT proteins and partially attenuated the effect of lncRNA TCL6 overexpression. TCL6 knockdown reduced PTEN protein level, further confirming that PTEN/PI3K/AKT signaling pathway was involved in lncRNA TCL6/miR-21 axis modulating Rb cell proliferation and apoptosis.
As a further confirmation of the above findings, miR-21 expression was significantly upregulated while PTEN mRNA expression was downregulated in Rb tissues. lncRNA TCL6 was negatively correlated with miR-21 while positively correlated with PTEN. Taken together, lncRNA TCL6 serves as a sponge for miR-21 to counteract miR-21-mediated repression of PTEN, therefore modulating Rb cell proliferation and apoptosis through PTEN/PI3K/AKT signaling pathways.
Figures and Tables
Fig. 1
Expression of long non-coding RNA T-cell leukemia/lymphoma 6 (lncRNA TCL6) in retinoblastoma (Rb) tissues and cell lines.
(A) Expression of lncRNA TCL6 in normal retina tissues (n = 8) and Rb tissue specimens (n = 22) were detected using real-time PCR. (B) Expression of lncRNA TCL6 in a normal cell line, ARPE-19, and two Rb cell lines, Y79 and WERI-Rb-1, were detected using real-time PCR. The data are presented as mean ± standard deviation of three independent experiments. *p < 0.05, **p < 0.01.

Fig. 2
Effect of long non-coding RNA T-cell leukemia/lymphoma 6 (lncRNA TCL6) knockdown on retinoblastoma (Rb) cell proliferation and apoptosis.
(A) LncRNA TCL6 knockdown was achieved in Y79 and WERI-Rb-1 cells by transfection of si-TCL6, compared to si-NC (negative control), as confirmed by real-time PCR. Y79 and WERI-Rb-1 cells were transfected with si-TCL6 and examined for cell viability using MTT assay (B), cell apoptosis using Flow cytometry (C), and DNA synthesis capacity using 5-ethynyl 2-deoxyuridine (EDU) assay (D). Apollo staining (red) and DAPI staining (blue) were performed to detect the EDU positive cells; The arrow indicated positive cells (×400). The data are presented as mean ± standard deviation of three independent experiments. **p < 0.01.
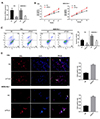
Fig. 3
miR-21 is a direct target of long non-coding RNA T-cell leukemia/lymphoma 6 (lncRNA TCL6).
(A, B) TCL6 overexpression (OE) vectors or miR-21 mimics were transfected into in Y79 and WERI-Rb-1 cells. The levels of TCL6 and miR-21 were determined using real-time PCR. (C) miR-21 expression in Y79 and WERI-Rb-1 cells examined using realtime PCR. (D, E) miR-21 expression in response to TCL6 knockdown or overexpression was detected in Y79 and WERIRb-1 cells. (F) The schematic diagram showing predicted miR-21 binding site in lncRNA TCL6. Two kinds of TCL6 luciferase reporter gene vectors containing wild or mutated miR-21 binding site were constructed. HEK293 cells were cotransfected with the above vectors and miR-21 mimics and examined for luciferase activity. NC, negative control; Wt, wild-type; mut, mutant-type. The data are presented as mean ± standard deviation of three independent experiments. **p < 0.01.
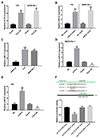
Fig. 4
The dynamic effect of long non-coding RNA T-cell leukemia/lymphoma 6 (lncRNA TCL6) and miR-21 on retinoblastoma (Rb) cell proliferation and apoptosis.
Y79 and WERI-Rb-1 cells were co-transfected with lncRNA TCL6 overexpressing vector (lncTCL6 OE) and miR-21 mimics, and examined for cell viability using MTT assay (A) and DNA synthesis capacity using 5-ethynyl 2-deoxyuridine (EDU) assay (B). The arrow showed the EdU positive cells. OD, optical density. The data are presented as mean ± standard deviation of three independent experiments. *p < 0.05, **p < 0.01 compared to vector + negative control (NC) mimics; #p < 0.05, ##p < 0.01, compared to TCL6 OE + NC mimics group.
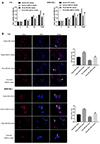
Fig. 5
PTEN/PI3K/AKT signaling pathway is involved in long non-coding RNA T-cell leukemia/lymphoma 6 (lncRNA TCL6)/miR-21 axis modulating retinoblastoma (Rb) cell proliferation.
(A) Y79 and WERI-Rb-1 cells were co-transfected with lncRNA TCL6 overexpressing (OE) vector and miR-21 mimics, and examined for protein levels of p-PTEN, PTEN, p-AKT, and AKT. (B) Y79 and WERI-Rb-1 cells were transfected with si-TCL6 and examined for protein level of p-PTEN, PTEN, p-AKT, and AKT. (C, D) The activity of PI3K was determined by ELISA. **p < 0.01 compared to vector + negative control (NC) mimics or NC group; #p < 0.05, ##p < 0.01, compared to TCL6 OE + NC mimics group.
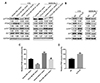
Fig. 6
Expression and correlation of miR-21 and PTEN in tissue specimens.
(A, B) Expression of miR-21 and PTEN mRNA expression in retinoblastoma (Rb) and non-cancerous tissue specimens was detected using real-time PCR. (C, D) Correlation between long non-coding RNA T-cell leukemia/lymphoma 6 (lncRNA TCL6) and miR-21, between lncRNA TCL6 and PTEN in Rb tissues was analyzed using Pearson's correlation analysis. The data are presented as mean ± standard deviation of three independent experiments. **p < 0.01, compared to the normal group.

Notes
References
1. Golabchi K, Soleimani-Jelodar R, Aghadoost N, Momeni F, Moridikia A, Nahand JS, Masoudifar A, Razmjoo H, Mirzaei H. MicroRNAs in retinoblastoma: potential diagnostic and therapeutic biomarkers. J Cell Physiol. 2018; 233:3016–3023.

2. Pérez-Ramírez C, Cañadas-Garre M, Molina MÁ, Faus-Dáder MJ, Calleja-Hernández MÁ. PTEN and PI3K/AKT in non-small-cell lung cancer. Pharmacogenomics. 2015; 16:1843–1862.

3. Chen H, Zhou L, Wu X, Li R, Wen J, Sha J, Wen X. The PI3K/AKT pathway in the pathogenesis of prostate cancer. Front Biosci (Landmark Ed). 2016; 21:1084–1091.
4. Gui F, Hong Z, You Z, Wu H, Zhang Y. MiR-21 inhibitor suppressed the progression of retinoblastoma via the modulation of PTEN/PI3K/AKT pathway. Cell Biol Int. 2016; 40:1294–1302.

5. Wei D, Miao Y, Yu L, Wang D, Wang Y. Downregulation of microRNA-198 suppresses cell proliferation and invasion in retinoblastoma by directly targeting PTEN. Mol Med Rep. 2018; 18:595–602.

6. Zou WW, Xu SP. Galangin inhibits the cell progression and induces cell apoptosis through activating PTEN and Caspase-3 pathways in retinoblastoma. Biomed Pharmacother. 2018; 97:851–863.

7. Xie C, Lu H, Nomura A, Hanse EA, Forster CL, Parker JB, Linden MA, Karasch C, Hallstrom TC. Co-deleting Pten with Rb in retinal progenitor cells in mice results in fully penetrant bilateral retinoblastomas. Mol Cancer. 2015; 14:93.

8. Su S, Gao J, Wang T, Wang J, Li H, Wang Z. Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in retinoblastoma. Tumour Biol. 2015; 36:7205–7211.

9. Hao F, Mou Y, Zhang L, Wang S, Yang Y. LncRNA AFAP1-AS1 is a prognostic biomarker and serves as oncogenic role in retinoblastoma. Biosci Rep. 2018; 38:BSR20180384.

10. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006; 103:2257–2261.

11. Abtin M, Alivand MR, Khaniani MS, Bastami M, Zaeifizadeh M, Derakhshan SM. Simultaneous downregulation of miR-21 and miR-155 through oleuropein for breast cancer prevention and therapy. J Cell Biochem. 2018; 119:7151–7165.

12. Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl). 2016; 7:19–27.
13. LArki P, Ahadi A, Zare A, Tarighi S, Zaheri M, Souri M, Zali MR, Ghaedi H, Omrani MD. Up-regulation of miR-21, miR-25, miR-93, and miR-106b in gastric cancer. Iran Biomed J. 2018; 22:367–373.

14. Shen F, Mo MH, Chen L, An S, Tan X, Fu Y, Rezaei K, Wang Z, Zhang L, Fu SW. MicroRNA-21 down-regulates Rb1 expression by targeting PDCD4 in retinoblastoma. J Cancer. 2014; 5:804–812.

15. Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018; 75:467–484.

16. Yang FY, Wang Y, Wu JG, Song SL, Huang G, Xi WM, Tan LL, Wang J, Cao Q. Analysis of long non-coding RNA expression profiles in clear cell renal cell carcinoma. Oncol Lett. 2017; 14:2757–2764.

17. Su H, Sun T, Wang H, Shi G, Zhang H, Sun F, Ye D. Decreased TCL6 expression is associated with poor prognosis in patients with clear cell renal cell carcinoma. Oncotarget. 2017; 8:5789–5799.

19. Gutschner T, Diederichs S. The hallmarks of cancer: a long noncoding RNA point of view. RNA Biol. 2012; 9:703–719.
20. Shang W, Yang Y, Zhang J, Wu Q. Long noncoding RNA BDNF-AS is a potential biomarker and regulates cancer development in human retinoblastoma. Biochem Biophys Res Commun. 2018; 497:1142–1148.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download