Streptococcus pneumoniae is a common pathogen that causes community-acquired infections, such as upper respiratory infections, pneumonia, meningitis, and sepsis.
1 Clinical syndromes are classified as noninvasive illnesses, such as contiguous spread from the nasopharynx or skin, and invasive illnesses causing infections in sterile body fluids or bacteremia.
2 Pneumococci are divided into serotypes based on capsular polysaccharide structures, and 93 serotypes are currently recognized. Serotype distribution varies by age, disease syndrome, and geography.
34 Ever since the licensure and routine administration of polysaccharide-protein pneumococcal conjugate vaccine (PCV), pneumococcal ecology has changed, and the serotype distribution in PCV era may be different.
5 Non-vaccine type (NVT) strains causing invasive pneumococcal disease (IPD) are circulating worldwide.
6 The vicious cycle of antibiotic exposure, selection of resistant organisms in the nasopharynx, and transmission of these organisms within the community, which leads to difficult-to-treat infections, has been interrupted to some extent by the introduction and routine use of PCV. Herein, we report an unvaccinated healthy woman with extensive IPD caused by multidrug-resistant (MDR)
S. pneumoniae of NVT suspected to have spread from a PCV13 vaccinated infant.
A 29-year-old healthy woman, who suffered from fever and headache for 3 days, was admitted to our hospital. She was diagnosed with frontal sinusitis and cranial epidural abscess (
Fig. 1).
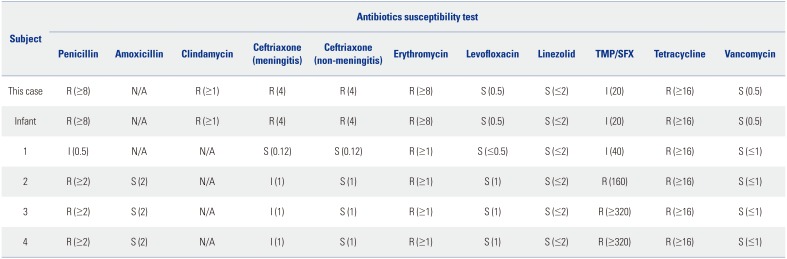
S. pneumoniae grew in the blood and tissue specimens from the frontal sinus obtained by a surgical approach. An antibiotic susceptibility test showed an MDR pathogen (
Table 1). Based on these results, the patient received levofloxacin (500 mg once daily) and vancomycin (1000 mg twice daily) for 2 weeks on admission and levofloxacin for 4 weeks in an outpatient clinic.
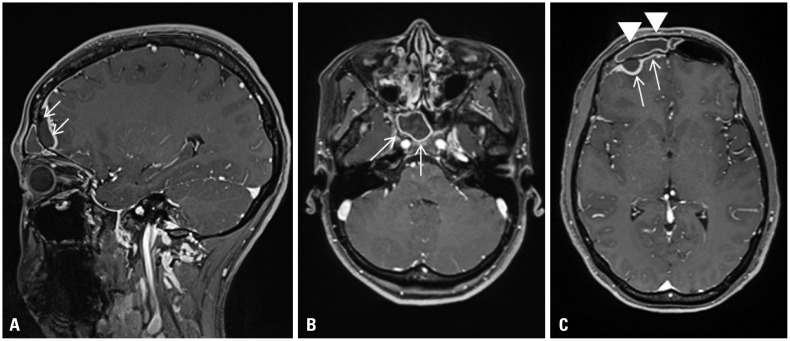 | Fig. 1Enhanced brain magnetic resonance imaging of 1.3×1.0×2.5 cm sized epidural empyema (A: arrows; C: arrows), and right frontal (C: arrowheads), ethmoid, and sphenoid sinusitis (B: arrows). Leptomeningeal enhancement with edema on right frontal convexity (C: arrows).
|
Table 1
Antibiotic Susceptibility Tests of Serotype 15B/C Streptococcus pneumoniae
|
Subject |
Antibiotics susceptibility test |
|
Penicillin |
Amoxicillin |
Clindamycin |
Ceftriaxone (meningitis) |
Ceftriaxone (non-meningitis) |
Erythromycin |
Levofloxacin |
Linezolid |
TMP/SFX |
Tetracycline |
Vancomycin |
|
This case |
R (≥8) |
N/A |
R (≥1) |
R (4) |
R (4) |
R (≥8) |
S (0.5) |
S (≤2) |
I (20) |
R (≥16) |
S (0.5) |
|
Infant |
R (≥8) |
N/A |
R (≥1) |
R (4) |
R (4) |
R (≥8) |
S (0.5) |
S (≤2) |
I (20) |
R (≥16) |
S (0.5) |
|
1 |
I (0.5) |
N/A |
N/A |
S (0.12) |
S (0.12) |
R (≥1) |
S (≤0.5) |
S (≤2) |
I (40) |
R (≥16) |
S (≤1) |
|
2 |
R (≥2) |
S (2) |
N/A |
I (1) |
S (1) |
R (≥1) |
S (1) |
S (≤2) |
R (160) |
R (≥16) |
S (≤1) |
|
3 |
R (≥2) |
S (2) |
N/A |
I (1) |
S (1) |
R (≥1) |
S (1) |
S (≤2) |
R (≥320) |
R (≥16) |
S (≤1) |
|
4 |
R (≥2) |
S (2) |
N/A |
I (1) |
S (1) |
R (≥1) |
S (1) |
S (≤2) |
R (≥320) |
R (≥16) |
S (≤1) |

We evaluated the patient's family members (her husband and 9-month-old infant) to detect the origin of the colonizer MDR
S. pneumoniae. Family members were subjected to nasopharyngeal (NP) swab culture as an outpatient procedure. The patient's blood, sputum, and tissue samples from inflammatory lesions of the frontal, sphenoid, and ethmoid sinus were examined. Specimens were streaked on 5% sheep blood agar plate, and then incubated for 24–48 hr at 37℃. In the case of blood, specimens were used after the positive signal in Bact/Alert 3D System (bioMerieux, Marcy-l'Etoile, France). For the identification of isolates, we used standard microbiological techniques using typical colonial appearance, hemolysis, Gram staining, bile solubility, susceptibility to optochin (1 µg) discs, and the automated VITEK II system (bioMerieux) with
S. pneumoniae ATCC 49619 for quality control. According to Clinical and Laboratory Standards Institute (CLSI) guidelines, separate interpretive breakpoints were used to define the resistance of isolates to each antimicrobial agent,
7 and an isolate resistant to three or more classes of antimicrobial agents was considered MDR. The study protocol was approved by the Institutional Review Board of Jeju National University Hospital (JNUH 19-02-007), and informed consent was obtained from all participants (the patient and family members).
S. pneumoniae isolates were re-inoculated onto blood agar plates to identify the molecular serotypes. Isolates with appropriate properties were serotyped by a sequetyping method, as described previously.
8 Genomic DNA was extracted from bacterial cells of a colony of pneumococci using heat lysis method. The primers used to amplify and sequence the cps gene were as follows:
cps1, 5′-GCA ATG CCA GAC AGT AAC CTC TAT-3′, and
cps2, 5′-CCT GCC TGC AAG TCT TGA TT-3′. The
cps locus was conserved in all serotypes, and the middle region of the operon contained serotype-specific genes that were used as targets of polymerase chain reaction (PCR)-based serotyping methods.
8 PCR amplicons were analyzed by 1.5% agarose gel electrophoresis. A second primer pair targeting the 16S rRNA gene was used to identify the species. Amplicons with the expected
cps band sizes (~1000 base pairs) were purified using a PCR Purification Kit, according to the manufacturer's instructions. Cycle sequencing was performed using BigDye Sequence Terminator v.3.1 (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's protocol. Amplicon nucleotide sequences were used to search against the GenBank database (
https://www.ncbi.nlm.nih.gov/nucleotide/), and the highest BLAST bit score (typically >98% identity) was used to identify the serotype. A phylogenetic tree was constructed based on capsular polysaccharide synthesis (
cps) gene sequences, using the maximum likelihood method implemented in MEGA 6.
9 The
cps sequence data for
S. pneumoniae were obtained from NCBI/BLAST (
https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The patient's infant was previously healthy, but had experienced an upper respiratory infection 15 days ago and took medicine from a local clinic. According to the Korean National Immunization Program (NIP), routine PCV 13 vaccination was administrated three times. The husband was also healthy, and did not show any sign and symptom of infection within the past three months.
S. pneumoniae was isolated from the NP swab culture of the patient's infant. However, the husband's culture was free of pathogens. When bacterial strain analysis was performed on samples from the patient and her infant, capsule serotype was 15B/C (strain 0556-97/3031-06; GenBank number, KY750640.1/KY750644.1). Antibiotic susceptibility test showed identical results for isolates of the mother and her infant (
Table 2), and their sequence types also matched (
Fig. 2).
 | Fig. 2Phylogenetic tree constructed based on capsular polysaccharide synthesis (cps) gene sequences. The cps sequences generated in this study are shown in bold (*Patient; †Patient's infant). Scale bar indicates nucleotide substitutions per site.
|
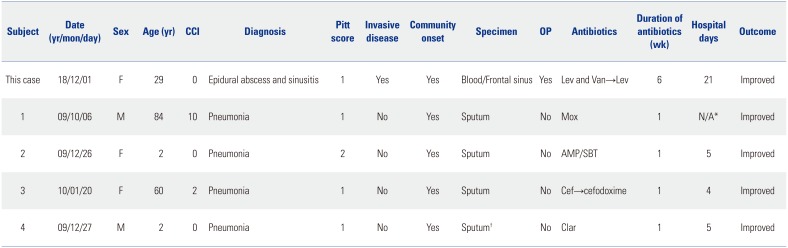
Table 2
Clinical Characteristics of Patients with Serotype 15B/C Streptococcus pneumoniae at Jeju National University Hospital
|
Subject |
Date (yr/mon/day) |
Sex |
Age (yr) |
CCI |
Diagnosis |
Pitt score |
Invasive disease |
Community onset |
Specimen |
OP |
Antibiotics |
Duration of antibiotics (wk) |
Hospital days |
Outcome |
|
This case |
18/12/01 |
F |
29 |
0 |
Epidural abscess and sinusitis |
1 |
Yes |
Yes |
Blood/Frontal sinus |
Yes |
Lev and Van→Lev |
6 |
21 |
Improved |
|
1 |
09/10/06 |
M |
84 |
10 |
Pneumonia |
1 |
No |
Yes |
Sputum |
No |
Mox |
1 |
N/A*
|
Improved |
|
2 |
09/12/26 |
F |
2 |
0 |
Pneumonia |
2 |
No |
Yes |
Sputum |
No |
AMP/SBT |
1 |
5 |
Improved |
|
3 |
10/01/20 |
F |
60 |
2 |
Pneumonia |
1 |
No |
Yes |
Sputum |
No |
Cef→cefodoxime |
1 |
4 |
Improved |
|
4 |
09/12/27 |
M |
2 |
0 |
Pneumonia |
1 |
No |
Yes |
Sputum†
|
No |
Clar |
1 |
5 |
Improved |

We report on the transmission of an MDR
S. pneumoniae of NVT causing cranial epidural abscess, sinusitis, and bacteremia from the nasopharynx of a PCV13-vaccinated infant to a young adult, and changes of antibiotics susceptibilities of serotype 15B/C in PCV era. Pneumococcal colonization is believed to represent the most important source of horizontal spread within the community.
10 Since children serve as major vectors of pneumococcal transmission, NP colonization may occur in adults via contact with children.
11 By 1 year of age, ~50% of children tend to experience at least one episode of pneumococcal colonization.
212 Although the introduction of PCV was followed by a successful reduction in IPD cases for both children and adults,
411 the frequency of PCV serotypes decreased while that of non-PCV serotypes increased among NP carriage pneumococci in the PCV13 era.
1113 A meta-analysis revealed that non-PCV13 serotypes contributed to 42.2% of childhood IPD cases, and non-PCV13 serotypes, such as 22F, 12F, 33F, 24F, 15B, 15C, 23B, 10A, and 38, were predominant in PCV era.
14 In addition, an increase in IPD caused by
S. pneumoniae of NVT has been reported.
1315 In particular, serotype 15B/C is currently among the most prevalent replacement for non-PCV13 serotypes,
16 and this serotype changes by capsular switching, which involves the emergence of NVT in a prevalent clone by replacement of the capsular polysaccharide locus, such as the switch from serotype 9V to 19A and from serotype 19A to 15B.
17
S. pneumoniae of the patient and her baby in this study were resistant to most antimicrobial agents tested, except linezolid, tigecycline, vancomycin, and levofloxacin, as observed by MDR patterns (
Table 1). Additionally, we investigated serotype 15B/C in our previous study.
13 Four serotypes 15B/C pneumococci were found in two children and two elderly patients (
Table 2). All patients had pneumonia. Serotype 15B/C of
S. pneumoniae isolates showed non-susceptibility to antibiotic classes (penicillins, macrolides, and tetracyclines) in 2009 and 2010. However,
S. pneumoniae isolated from this patient and her infant showed resistance to antibiotic classes (penicillins, lincosamides, macrolides, cephalosporin, and tetracyclines) in 2018. Compared to 2009 and 2010, the resistance pattern for
S. pneumoniae of 15B/C changed within the community in 2018. It is important to monitor the frequency of serotype exchanges in order to predict the long-term efficacy of PCV vaccines.
Our study had some limitations. First, we could not definitely conclude that the same serotype had spread from the infant to the mother, since it may have been transmitted from another patient or colonizer with S. pneumoniae of 15B/C serotype. Second, our data may not reflect the nationwide serotype and clonal distributions.
IPD of NVT can spread among healthy adults by close contact between family members. Continuous monitoring of antimicrobial resistance and serotype distribution of S. pneumoniae is important for disease management in the Korean population. Further surveillance and studies of serotype distribution in the community are necessary in the PCV 13 era.








 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download