Abstract
Canine adenovirus type 1 (CAV-1) infection results in hepatitis in dogs. In this study, we investigated the biologic and genetic characteristics of the CAV-1 vaccine strain (CAV1V) to improve quality control about CAV vaccine. The identity of CAV1V as CAV-1 was confirmed based on its cytopathic effects and the results of hemagglutination (HA) and immunofluorescence assays, and electron microscopy. The CAV1V strain reached 107.5 TCID50/mL in MDCK cells at 4 days post-inoculation and exhibited hemmagglutination activity of 256 U using guinea pig erythrocytes. Intranuclear fluorescence in the infected cells was observed and typical adenoviruses were observed in electon microscope. CAV1V strain was identified as a CAV-1 strain by nucleotide sequence analysis. In a comparison of the nucleotide sequences of the fiber genes of several CAV strains, CAV1V showed the highest similarity (99.8%) with the GLAXO strain, which was isolated in Canada. Our biological characterization of CAV1V will facilitate quality control of the canine hepatitis vaccine.
Go to : 
REFERENCES
1). Hu RL, Huang G, Qiu W, Zhong ZH, Xia XZ, Yin Z. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet Res Commun. 2001; 25:77–84.
2). Linné T. Differences in the E3 regions of the canine adenovirus type 1 and type 2. Virus Res. 1992; 23:119–33.

3). Walker D, Abbondati E, Cox AL, Mitchell GB, Pizzi R, Sharp CP, et al. Infectious canine hepatitis in red foxes (Vulpes vulpes) in wildlife rescue centres in the UK. Vet Rec. 2016; 178:421.

4). Yang DK, Kim HH, Yoon SS, Lee H, Cho IS. Isolation and identification of canine adenovirus type 2 from a naturally infected dog in Korea. Korean J Vet Res. 2018; 58:177–82.

5). Aoki E, Soma T, Yokoyama M, Matsubayashi M, Sasai K. Surveillance for Antibodies against Six Canine Viruses in Wild Raccoons (Procyon Lotor) in Japan. J Wildl Dis. 2017; 53:761–8.

6). Yang DK, Kim HH, Yoon SS, Ji M, Cho IS. Incidence and Sero-survey of Canine Adenovirus Type 2 in Various Animal Species. J Bacteriol Virol. 2018; 48:102–8.

7). Loustalot F, Kremer EJ, Salinas S. The Intracellular Domain of the Coxsackievirus and Adenovirus Receptor Differentially Influences Adenovirus Entry. J Virol. 2015; 89:9417–26.

8). Tham KM, Horner GW, Hunter R. Isolation and identification of canine adenovirus type-2 from the upper respiratory tract of a dog. N Z Vet J. 1998; 46:102–5.

9). Shenk TE. Adenoviridae: The Viruses and Their Replication. Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 4th ed. 2. Philadelphia: Lippincott Williams & Wilkins;2001. p. 2265–300.
10). Walker D, Fee SA, Hartley G, Learmount J, O'Hagan MJ, Meredith AL, et al. Serological and molecular epidemiology of canine adenovirus type 1 in red foxes (Vulpes vulpes) in the United Kingdom. Sci Rep. 2016; 6:36051.

11). Whetstone CA. Monoclonal antibodies to canine adenoviruses 1 and 2 that are type-specific by virus neutralization and immunofluorescence. Vet Microbiol. 1988; 16:1–8.

12). Morrison MD, Onions DE, Nicolson L. Complete DNA sequence of canine adenovirus type 1. J Gen Virol. 1997; 78:873–8.

13). Pizzurro F, Marcacci M, Zaccaria G, Orsini M, Cito F, Rosamilia A, et al. Genome Sequence of Canine Adenovirus Type 1 Isolated from a Wolf (Canis lupus) in Southern Italy. Genome Announc. 2017; 5.

14). Green NM, Wrigley NG, Russell WC, Martin SR, McLachlan AD. Evidence for a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J. 1983; 2:1357–65.

15). Mei YF, Wadell G. Hemagglutination properties and nucleotide sequence analysis of the fiber gene of adenovirus genome types 11p and 11a. Virology. 1993; 194:453–62.

16). Reed LJ, Muench H. A Simple Method of Estimating Fifty Per Cent Endpoints. Am J Epidemiology. 1938; 27:493–7.
17). Choi JW, Lee HK, Kim SH, Kim YH, Lee KK, Lee MH, et al. Canine adenovirus type 1 in a fennec fox (Vulpes zerda). J Zoo Wildl Med. 2014; 45:947–50.

18). Wong M, Woolford L, Hasan NH, Hemmatzadeh F. A Novel Recombinant Canine Adenovirus Type 1 Detected from Acute Lethal Cases of Infectious Canine Hepatitis. Viral Immunol. 2017; 30:258–63.

19). Takamura K, Ajiki M, Hiramatsu K, Takemitsu S, Nakai M, Sasaki N. Isolation and properties of adenovirus from canine respiratory tract. Nihon Juigaku Zasshi. 1982; 44:355–7.

20). Kennedy FA, Mullaney TP. Disseminated adenovirus infection in a cat. J Vet Diagn Invest. 1993; 5:273–6.

21). Chaturvedi U, Tiwari AK, Ratta B, Ravindra PV, Rajawat YS, Palia SK, et al. Detection of canine adenoviral infections in urine and faeces by the polymerase chain reaction. J Virol Methods. 2008; 149:260–3.

22). Sanjuá n R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016; 73:4433–48.
Go to : 
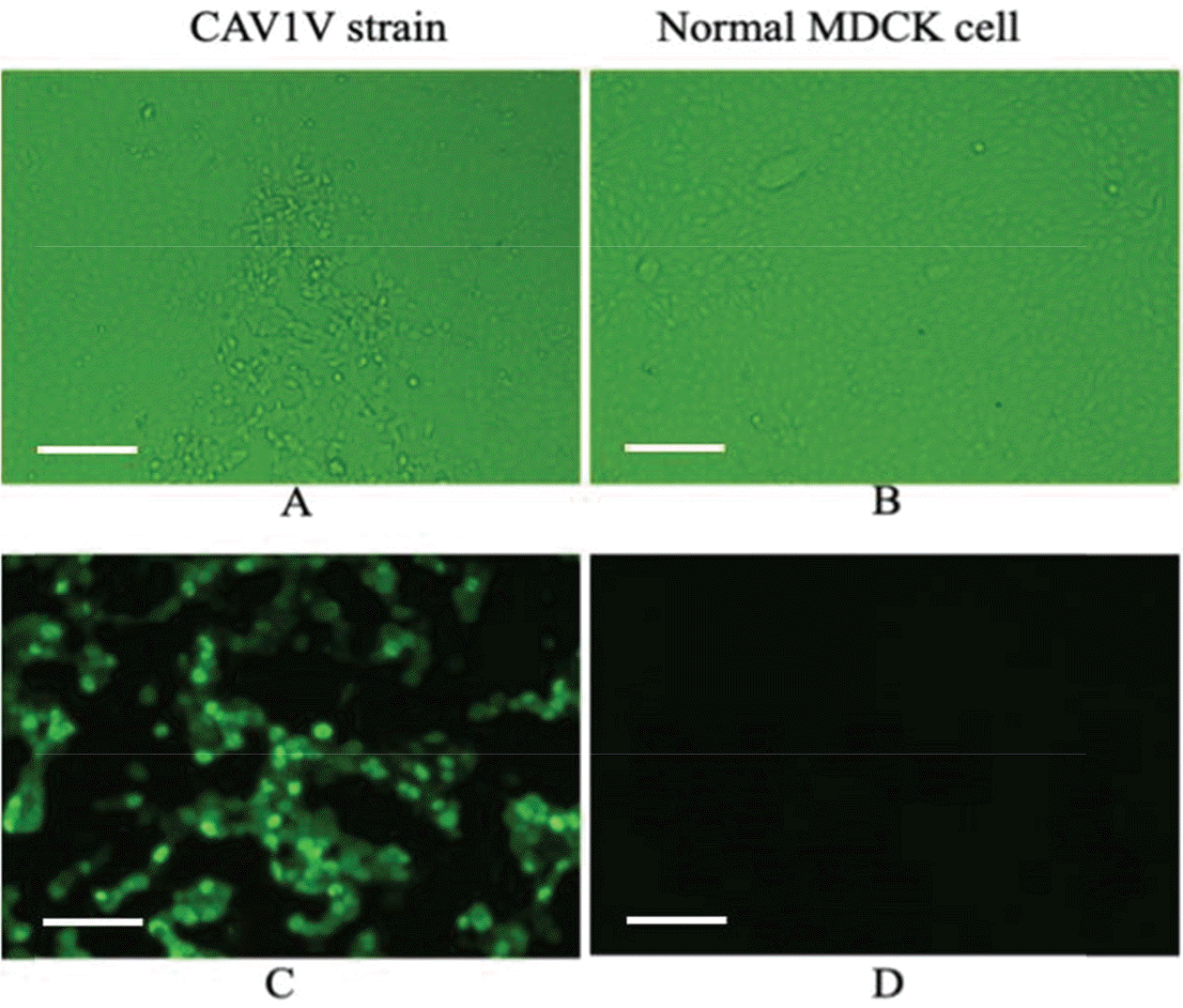 | Figure 1.Cytophatic effects in MDCK cells infected with CAV1V (magnification, 200×) (A). Normal MDCK cells derived from kidney of an adult female cocker spaniel (B). Indirect IF using a monoclonal antibody against CAV-1 (C, D). Intranuclear fluorescence was observed in MDCK cells infected with CAV1V (C) but not in normal MDCK cells (D). Scale bars, 100 μm (A-D). |
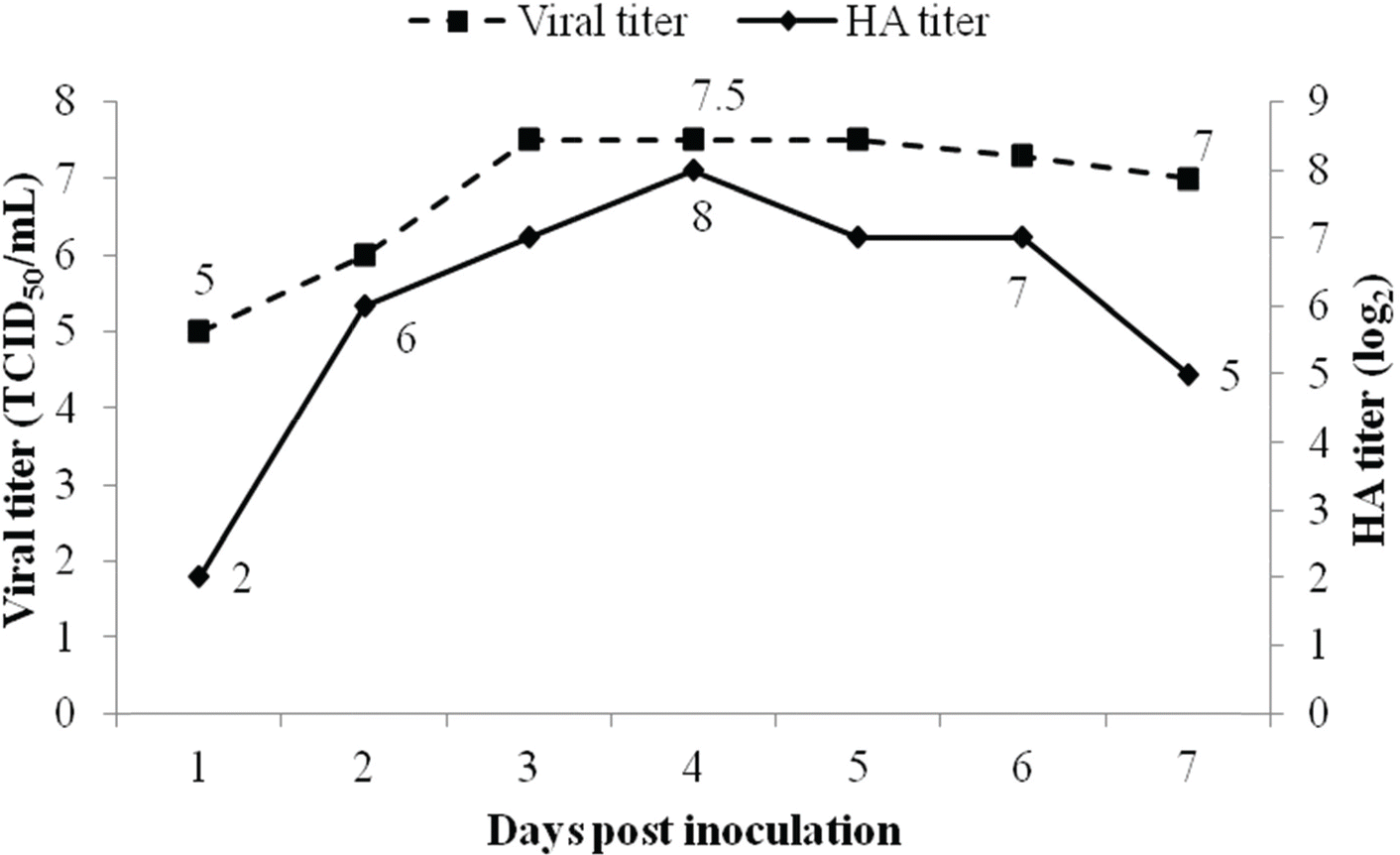 | Figure 2.Growth kinetics of CAV1V in MDCK cells according to time of harvest. At 4 days post inoculation, CAV1V strain was present at 107.5 TCID50/mL and 256 HA U/50 μL. |
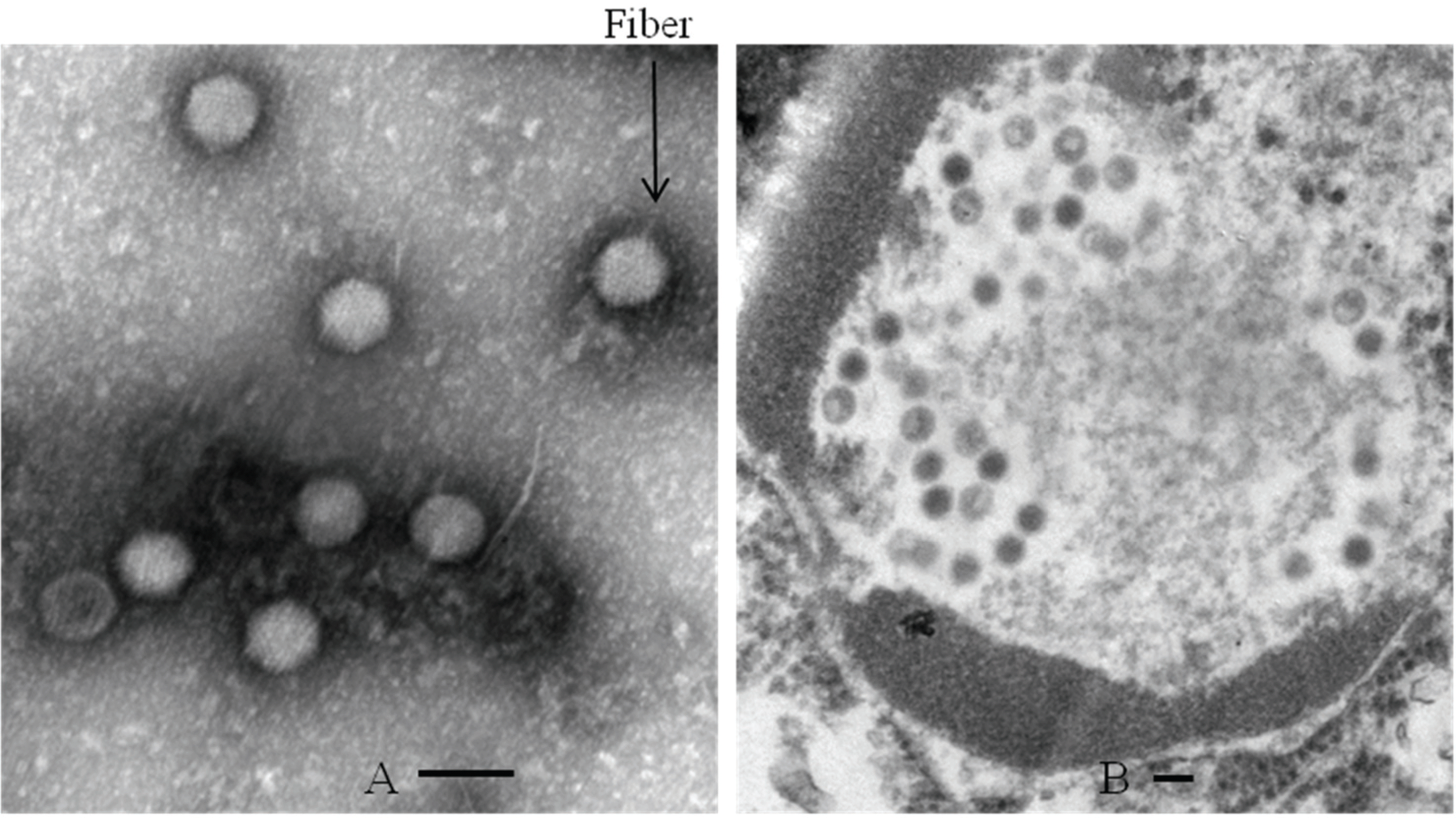 | Figure 3.Electron microscopic images of virus particles of 70–90 nm diameter. Fiber proteins of CAV1V purified with cesicum chloride gradient are protruding from the virion surface (A) (magnification, 100,000×). CAV particles are shown in the nucleus of MDCK celle (magnification, 50,000×) (B). Scale bars, 100 nm (A and B). |
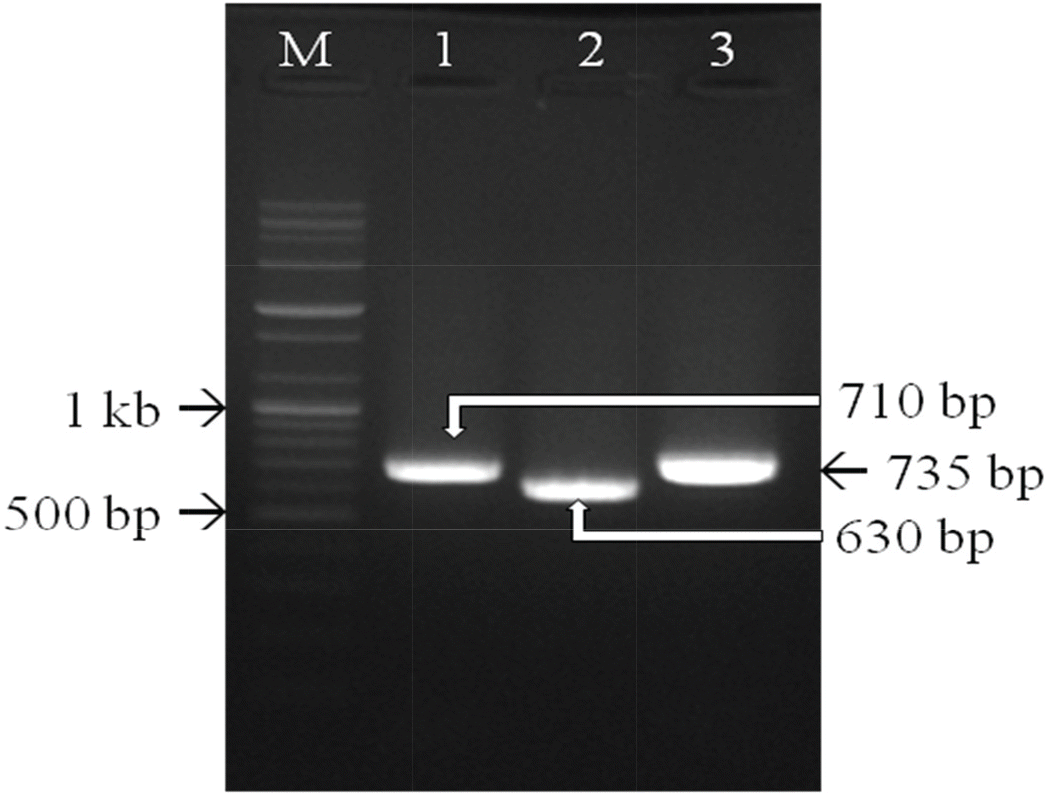 | Figure 4.Three primer sets targeting the fiber gene of CAV-1 were used for PCR. PCR products of the expected sizes confirmed that CAV1V was a CAV-1 strain. Lane M, 1 kb ladder; lanes 1-3, DNA amplified from CAV1V. |
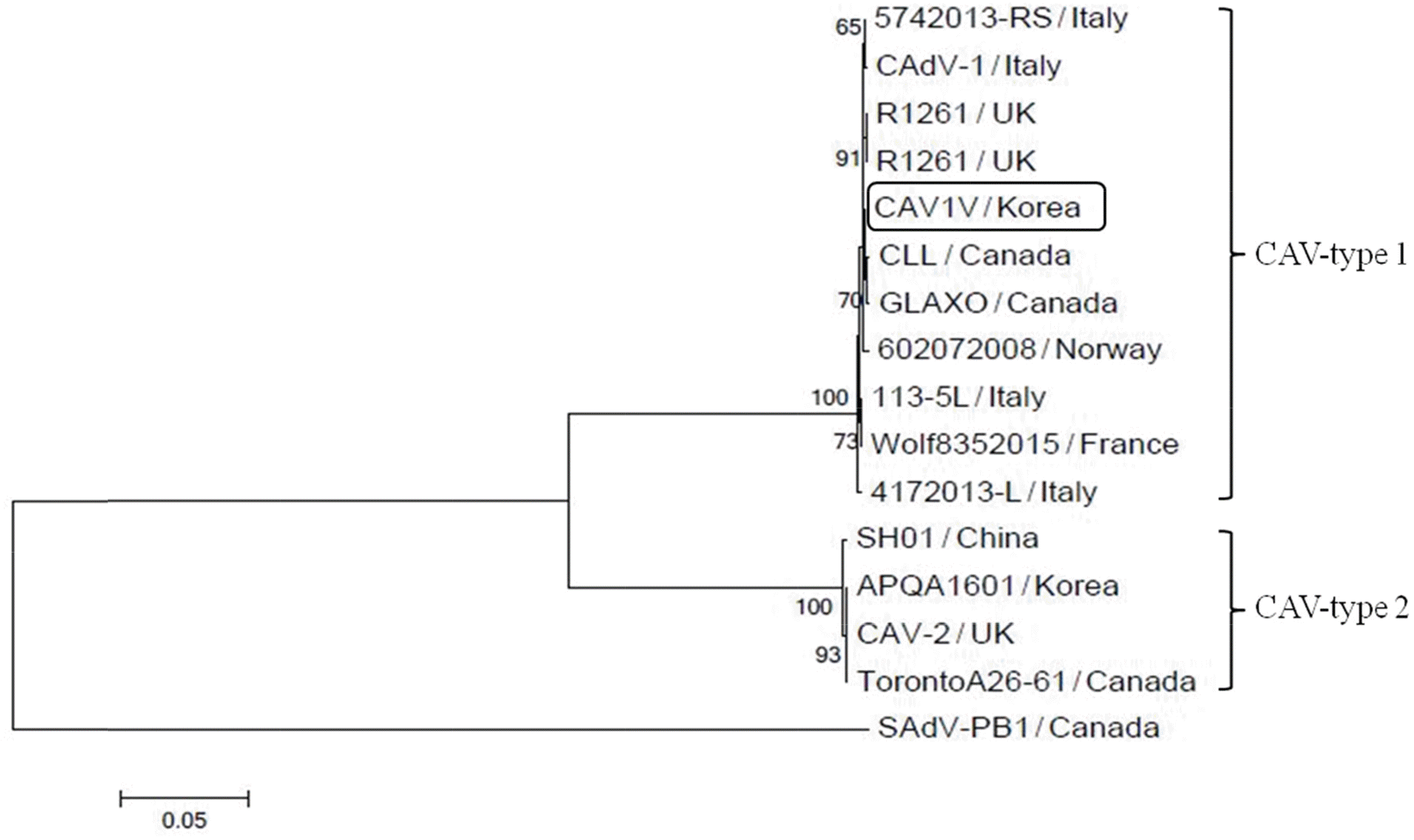 | Figure 5.Phylogenetic tree based on the nucleotide sequences of the fiber genes of 16 adenoviral strains. CAV1V belonged to the CAV-1 clade and had the highest homology with the GLAXO strain, which was isolated in Canada. The phylogenetic tree was constructed based on alignments of nucleotide sequences obtained using the neighbor-joining method. |
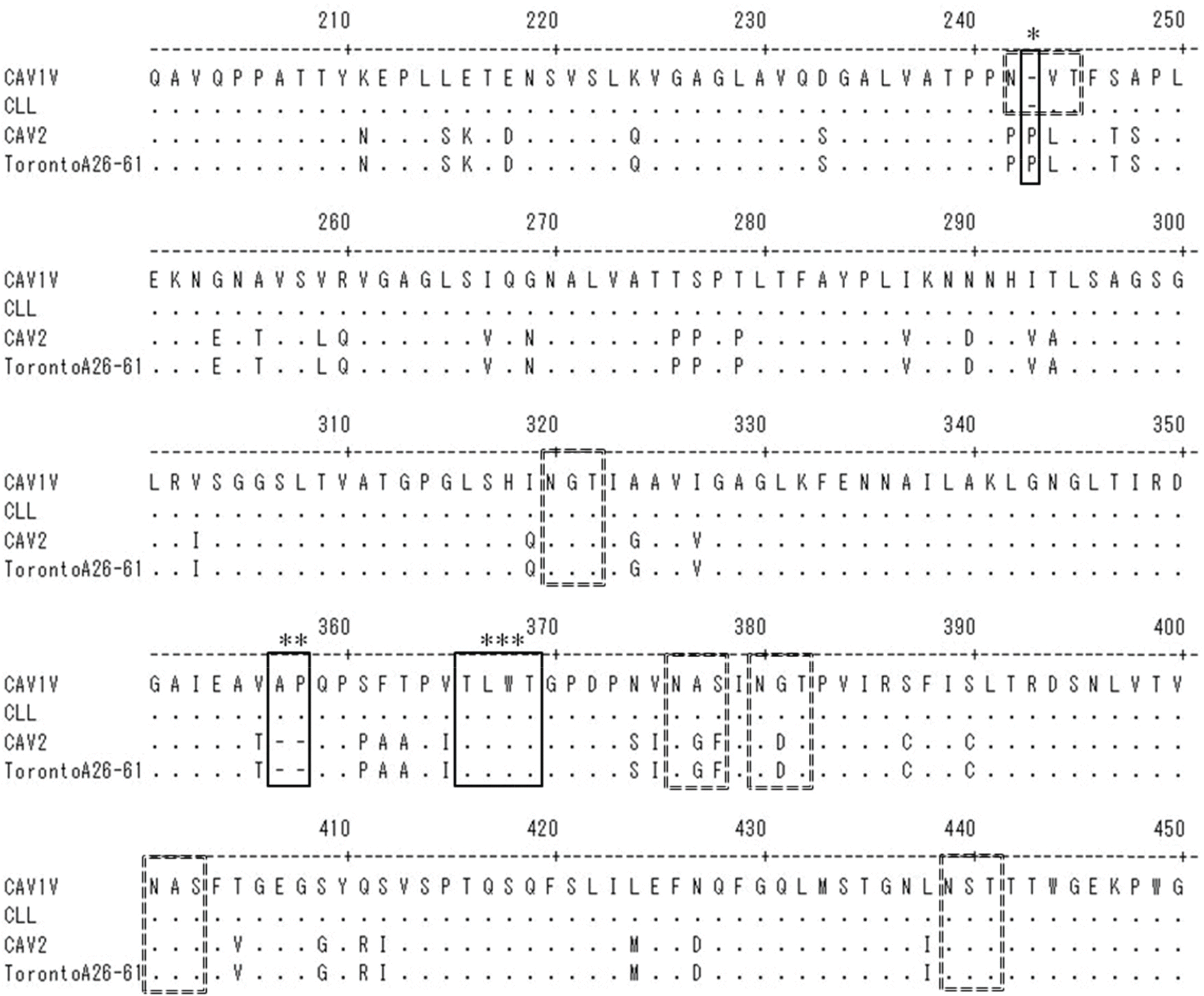 | Figure 6.Alignment of the predicted amino acids at positions 200-450 of the CAV1V fiber protein with the corresponding region of the fiber proteins of three other CAV strains. Dots indicate identical amino acids. Compared with CAV-2, there is one deletion (∗) and one insertion (∗∗) in the fiber protein of CAV1V. The region (∗∗∗) conserved in adenoviruses of all animal species was well preserved. Six potential N-linked glycosylation sites (N-X-S/T) are indicated by dotted-line boxes. |
Table 1.
The primers used for the PCR-based analysis of canine adenovirus type 1




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download