Abstract
Objective
This study aimed to investigate the effect of pre-applied orthodontic force on the regeneration of periodontal ligament (PDL) tissues and the underlying mechanisms in tooth replantation.
Methods
Orthodontic force (50 cN) was applied to the left maxillary first molars of 7-week-old male Sprague–Dawley rats (n = 32); the right maxillary first molars were left untreated to serve as the control group. After 7 days, the first molars on both sides were fully luxated and were immediately replanted in their original sockets. To verify the effects of the pre-applied orthodontic force, we assessed gene expression by using microarray analysis and real-time reverse transcription polymerase chain reaction (RT-PCR), cell proliferation by using proliferating cell nuclear antigen (PCNA) immunofluorescence staining, and morphological changes by using histological analysis.
Results
Application of orthodontic force for 7 days led to the proliferation of PDL tissues, as verified on microarray analysis and PCNA staining. Histological analysis after replantation revealed less root resorption, a better arrangement of PDL fibers, and earlier regeneration of periodontal tissues in the experimental group than in the control group. For the key genes involved in periodontal tissue remodeling, including CXCL2, CCL4, CCL7, MMP3, PCNA, OPG, and RUNX2, quantitative RT-PCR confirmed that messenger RNA levels were higher at 1 or 2 weeks in the experimental group.
The management of missing teeth in both adolescents and adults can be quite challenging. Implant prostheses have been widely used to successfully replace the missing teeth with predictable esthetic results. These prostheses, however, have several disadvantages, such as the lack of physiological proprioception and the absence of sensory response to chewing provided by the periodontal ligament (PDL). Furthermore, implants do not allow the vertical bone growth that occurs in the natural teeth of growing patients, leading to differential gingival levels.1 Recently, autotransplantation of teeth has been attempted, with high rates of success (approximately 90%) being reported in previous study.2 Tooth autotransplantation is cost-effective in that it enables the transfer of nonfunctional teeth (usually third molars) to a functional position. In addition, in young patients who are still growing, a transplanted tooth with healthy PDL tissue can erupt in harmony with the adjacent teeth. For these reasons, autotransplantation is suggested to be the treatment of choice when tooth rehabilitation is needed.
Various forms of management have been proposed for tooth transplantation,3 and the responses of the periodontal tissue surrounding transplanted teeth have been well described.4 One common complication of autotransplantation is ankyloses, which is associated with root resorption. Therefore, for autotransplantation to be successful, it is essential to minimize the incidence of root resorption and ankylosis. Previous study has shown that to accomplish this goal, periodontal healing along with preservation of the remaining PDL tissues and cementum is an important step.5
Studies have suggested that the application of orthodontic force leads to the differentiation and proliferation of PDL tissues through increases in vascularity6 and in the recruitment of osteoblast precursors,7 as well as changes in the expression levels of various chemokines.8 Moreover, PDL tissue proliferation resulted in widening of the PDL space and easier extraction. Since preservation of PDL tissues is key to successful autotransplantation, and these tissues can proliferate or become activated in response to orthodontic force, we preconditioned PDL tissue by applying orthodontic force to determine its effects on the regeneration of periodontal tissues.
We obtained 32 Sprague–Dawley rats that were 7 weeks old, comparable with adolescence in humans, and weighed approximately 170 g. All animal subjects were housed in animal care–approved laboratory facilities and were fed a solid diet and water ad libitum. The entire procedure met the ethical standards issued by the Animal Care and Use Committee of Chonnam National University (No. CNU IACUC-YB-2017-51).
A pilot study was carried out prior to the main study to verify changes in the PDL as a result of applying orthodontic force for 1 week. Figure 1 illustrates the two study designs.
After being briefly anesthetized by inhalation of diethyl ether, the rats were fully anesthetized by intraperitoneal injection of ketamine (50 mg/kg). To stimulate the PDL tissues prior to autotransplantation, orthodontic force was applied to the left maxillary first molars (the experimental group; n = 32), while the right maxillary first molars were left untreated (the control group; n = 32). Superelastic nickel-titanium (NiTi) closed-coil springs (Tomy International, Yokohama, Japan) were used for tooth movement and were ligated between the left maxillary first molar and the maxillary incisors. The NiTi coil spring was fixed with 0.009-inch stainless-steel wire, and a force of 50 cN was applied with calibration by using a tension gauge (Federwaage, Gersbach, Germany). A shallow groove was made on the labial surface of the incisors, and stainless-steel wire was bonded to this groove by using light-cured adhesive and resin. The mandibular incisor edge was trimmed to prevent detachment of the appliance from the occlusion (Supplementary Figure 1). The appliance was maintained for 7 days before luxation of the first molars. Both right and left maxillary first molars were subluxated buccopalatally until fully luxated.
The same procedures were carried out for the control molars. To stabilize the replanted molars, semi-rigid splinting of the first and second molars was performed using 0.009-inch ligature wire and resin application (Supplementary Figure 2). Eight rats in each group were sacrificed at 1, 2, 3, and 4 weeks after replantation, and the tissues were analyzed for histological changes (n = 3) and messenger RNA (mRNA) analysis (n = 5).
To investigate molecular changes in the PDL tissue, right- and left-side tissue blocks containing the only first molar roots were extracted 1 week after the application of orthodontic force. Total RNA was isolated using TRIzol Reagent (Molecular Research Center, Cincinnati, OH, USA) and was quantified using spectrophotometry. Profiling of gene expression was performed using Agilent 22K 60-mer oligonucleotide microarray slides (Digital Genomics, Seoul, Korea), which contained approximately 22,000 distinct sequences. The fluorescent-labeled complementary RNA (cRNA) was prepared by amplification of the total RNA in the presence of deoxyuridine triphosphate (dUTP), followed by the coupling of Cy3 (experiment) and Cy5 (control) dyes (Amersham-Pharmacia, Uppsala, Sweden). An Oligo Microarray Kit was hybridized with the fluorescent-labeled cRNA at 60℃ for 16 hours. The DNA chips were scanned using GenePix 4000B (Axon Instruments, Union City, CA, USA), and the scanned images were analyzed using GenePix Pro 3.0 software (Axon Instruments) to obtain the gene expression ratios. The logged gene expression ratios were normalized by LOWESS (locally weighted scatter plot smoother) regression.9
To evaluate histological changes at 1, 2, 3, and 4 weeks after replantation, three rats in each group were sacrificed by transcardiac perfusion of 4% paraformaldehyde solution (pH 7.4). The specimens containing the left and right maxillary molar regions were removed for fixation in the same solution of 4% paraformaldehyde, decalcified in ethylenediaminetetraacetic acid (EDTA) solution (pH 7.4), and routinely processed for embedding in paraffin. The serial sagittal sections, 5 µm in thickness, were prepared and stained with hematoxylin-eosin (Sigma-Aldrich Co., St. Louis, MO, USA) for morphological analysis.
To verify proliferation of the PDL tissue as a result of the orthodontic force, we used rat polygonal primary antibodies for proliferating cell nuclear antigen (PCNA), and immunofluorescence staining was performed using a tyramide signal amplification (TSA) kit (Invitrogen, Carlsbad, CA, USA). The 5-µm-thick sections were obtained from the tissues of both the experimental and the control groups. Each section was placed on a slide, fixed with 4% paraformaldehyde for 20 minutes, and then washed with phosphate-buffered saline for 5 minutes. We used 3% H2O2 to block endogenous peroxidase for 1 hour, and the sections were reacted with the primary antibody for 1 day, followed by reaction with biotinylated secondary antibody for 1 hour. Tyramide working solution was used to colorize the reactants. The reactants were visualized and photographed using a confocal laser scanning microscope (LSM) (Carl Zeiss, Göttingen, Germany). Immunological specificity was assessed by substituting the primary antibody from the normal serum.
To evaluate the changes in gene expression after replantation, five rats from each group were sacrificed by an overdose of CO2, and total RNA was extracted using TRIzol Reagent (Gibco BRL, Gaithersburg, MD, USA). The RNA was stored at −70℃ and was measured at an optical density of 260 nm. The mixtures of total RNA were incubated with Oligo dT (Gibco BRL), and a complementary DNA (cDNA) synthesis reaction was performed using a mixture of AccuPower PCR PreMix (Bioneer, Daejeon, Korea) and water. The cDNA was synthesized from the total RNA obtained from both groups by using the SuperScript First-Strand Synthesis System (Invitrogen) and was amplified with a Rotor-Gene RG-3000 (Corbett Research, Mortlake, VIC, Australia). The amplified cDNA was detected with the SYBR Green PCR Master Mix Reagent Kit (Qiagen, Germantown, MD, USA).
PCR conditions were as follows: incubation for 10 minutes at 95℃, followed by 45 cycles of 10-seconds denaturation at 95℃, annealing for 15 seconds at 60℃, and 20-seconds extension at 72℃. A reaction mixture lacking cDNA was used as the negative control in each run. Primer sequences are summarized in Supplementary Table 1. Data were analyzed using the Rotor-Gene 6 (Corbett Robotics, Brisbane, QLD, Australia). Ratios of the intensities of the target genes and β-actin signals were used as a relative measure of the expression levels of the target genes. To ensure accuracy of the experiment, real-time PCR assays were performed in triplicate for each sample. The mean fold change in the expression levels in the experimental group in comparison with the control group was calculated from the comparative Ct (ΔΔCt) values, and the range of the fold changes was represented by the standard deviations of the values.10
We used microarray analysis to compare the gene expression profile of PDL tissue after the application of orthodontic force with that of the control PDL tissue. The global M-values (log2 [R/G]; R = Cy5 signal:background, G = Cy3 signal:background) of the intensity of each spot were calculated after global normalization. Expression rates of 30,367 genes were investigated. Among them, 2,520 genes showed more than 2-fold fewer changes in expression; 1,092 genes (43%) were upregulated, including CXCL2 (macrophage inflammatory protein [MIP]-2α, 16.4-fold), CCL4 (MIP-1β, 10.2-fold), CCL7 (monocyte chemotactic protein-3 [MCP3], 8.0-fold), and MMP3 (matrix metalloproteinase 3, 22.4-fold), whereas 1,428 (57%) were downregulated (Supplementary Table 2). Supplementary Figure 3 shows the cluster analysis of gene changes.
Supplementary data is available at https://doi.org/10.1041/kjod.2019.49.5.299.
To detect cell arrangement and behavior, tissue sections from the samples were stained with hematoxylin and eosin. In the control group without force, the PDL space was relatively even, and the surface of the alveolar bone facing the PDL was smooth. In the orthodontic force group, PDL tissues on the tension side appeared tensed and the cells were arranged horizontally and widened, whereas the tissues on the pressure side were compressed and narrowed, with the cells arranged vertically (Figure 2A).
PCNA immunofluorescence staining was performed to validate PDL cell proliferation. The reactivity in the control group (without orthodontic force) was relatively sparse and weak. In contrast, strong immunoreactivity and an increased number of reactive cells were observed not only in the PDL but also in the alveolar bone of the experimental group (Figure 2B).
In the control group, the direction and arrangement of the PDL tissue around the root surface was more irregular, and cementogenesis with cellular cementum occurred around the apex of the root. Many osteoclasts were observed on the surface of alveolar bone, and localized root resorption was frequently seen at the root surface, as compared with the experimental group. Enlarged blood vessels with nearby inflammatory cells were notable in the control group (Figure 3A). In the experimental group, the PDL tissues were relatively more oriented, as can be seen in the oblique fibers, and mild angiogenesis was observed (Figure 3B).
Localized inflammatory reactions and root resorption were still evident in the control group (Figure 3C). In the experimental group, the direction of PDL collagen fibers became more regular, and blood vessels were normally localized at the bony side rather than at the cementum side. Parts of the PDL tissues appeared to have matured (Figure 3D).
The regeneration of the PDL in the control group was ameliorated in that the PDL cells were obliquely oriented and many blood vessels were organized near the alveolar bone. However, localized root resorption, particularly in the apical region, and infiltration of inflammatory cells were still evident (Figure 3E). In the experimental group, the periodontal tissues showed normal figures, but rearrangement of PDL cells and localized cementum regeneration could still be seen (Figure 3F).
In the control group, the PDL tissue showed normally arranged figures and inflammatory reactions were hardly seen. However, localized resorption of the root in the apical region was being recovered by cellular cementum regeneration (Figure 3G). In the experimental group, the periodontal tissues and roots showed normal appearances (Figure 3H).
In order to validate the expression levels of the genes analyzed by microarray, several key genes involved in periodontal tissue remodeling, including chemokines (CXCL2, CCL4, and CCL7) and MMP3, were further studied by means of real-time RT-PCR. Also investigated were the genes for osteoprotegerin (OPG) and runt-related transcription factor 2 (RUNX2), which are involved in osteoclastogenesis and osteoblastogenesis, respectively, and PCNA, a marker of cell proliferation. The time-course expression of these genes of interest is shown in Figure 4. In comparison with the control group, CXCL2 mRNA was upregulated 10.0-fold at 1 week (p < 0.001) and 2.5-fold at 2 weeks (p < 0.05). Similarly, CCL4 mRNA was upregulated 5.5-fold at 1 week (p < 0.001), whereas CCL7 mRNA was maintained at its control level until 2 weeks, followed by upregulation at 3 weeks after replantation. The transcription level of MMP3, a matrix-degrading enzyme, was upregulated 8.0-fold at 1 week (p < 0.001) and 3.0-fold at 2 weeks (p < 0.001), followed by downregulation to the control level. At 1 week, OPG mRNA (2.0-fold, p < 0.05), RUNX2 mRNA (8.0-fold, p < 0.001), and PCNA mRNA (3.0-fold, p < 0.01) were upregulated and then returned to control levels.
Autotransplantation has been widely performed since it provides patients with all the benefits of natural teeth. However, autotransplantation can fail if ankylosis and root resorption occur during the periodontal tissue healing process.11
Previous studies have reported that successful transplantation depends on four factors that reduce the occurrence of ankylosis and any type of root resorption: a) the condition of the remaining PDL tissue attached to the extracted donor tooth,12 b) accurate adaptation of the donor tooth to the recipient site,13 c) the duration and method of splinting after transplantation,214 and d) the schedule for endodontic treatment of the transplanted teeth.15 All but the first of these factors could be influenced by the clinician's technique. Mechanical stimuli can cause the PDL tissues of the donor tooth to proliferate and become activated, and these tissues often remain around the root surface after the tooth has been extracted. Katayama et al.16 have reported the effect of such proliferation on transplanted teeth. They transplanted proliferation tissue at a recipient site during tooth transplantation and suggested that this tissue might promote regeneration of the periodontal tissue and prevent dentoalveolar ankylosis and root resorption. However, their approach differed from this study in that they used tissue acquired from the healing site 2 weeks after extraction. Therefore, these transplanted tissues not only were different from the PDL cells in their cell arrangement but also were not activated. Thus, rather than adding inert tissue to the site of transplantation, we investigated the effect of direct activation of in vivo PDL tissues attached to the root surface upon autotransplantation.
The appliance design, amount of force, and period of orthodontic force application were determined according to a previous study1718 in which the authors systematically evaluated the force delivery system and experimental models to detect tooth movement and proposed appropriate experimental models in rats. Ideally, the optimal orthodontic force for moving molars in the rat is less than 20 cN, which is equivalent to a force of 400 cN in humans. However, in most of the previous studies, 50 cN was a practical choice owing to the difficulty in handling the orthodontic appliances. Since simulation of autotransplantation in the rat is a challenging process, and replantation of teeth has been the acceptable alternative experimental model, we used the replantation model described in a previous study by Suzaki et al.19 These authors subluxated the tooth buccopalatally until the mesiobuccal root was dislocated 1 mm and then replanted the tooth immediately in the same place. In this study, however, the tooth was fully luxated without any trauma and replanted in the same place without any delay.
Because the occlusal force has been reported to promote periodontal healing,14 the replanted tooth was fixed using the semi-rigid fixation method to allow biological tooth movement during the periodontal healing process.
In this study, the healing process in the control group showed many characteristic features, such as an irregular arrangement of cells in the PDL tissue and evidence of root resorption. However, there was a major difference in healing features between the control and experimental groups at 1 week, and healing continued to be delayed in the control group for up to 4 weeks. Even though the presence of inflammatory cells and irregularly arranged PDL tissues did not persist over time in the control group, healing was slower when compared with that in the experimental group during the entire healing process. This result was similar to the findings of a previous study by Katayama et al.16 and suggested that proliferated and activated PDL tissue promoted the reconstruction of PDL tissue after transplantation. Activated PDL tissues may secrete molecules, including chemotactic factors and vascular endothelial growth factor, to induce the formation of blood vessels and mesenchymal stem cells to regenerate periodontal tissue.
Recent studies have suggested that the chemokines provide the essential signal for the mobilization and recruitment of osteoblast and osteoclast precursors, which are also essential for the development, activity, and survival of bone cells.20 Therefore, chemokines are considered to be the potential modulators of bone remodeling and tissue regeneration.21 In this study, seven genes of interest were selected to study the time-course expression after replantation in both the experimental and control groups. Based on the results of microarray analysis, we chose four genes with the most upregulated mRNA (CXCL2, CCL4, CCL7, and MMP3) and two more genes (OPG and RUNX2) were selected based on current scientific knowledge. PCNA mRNA was also investigated to verify whether PDL tissue would proliferate in response to the pre-application of orthodontic force.
CXCL2 is the chemokine induced by the orthodontic force, especially pressure, resulting in inflammatory reactions and bone remodeling.22 CXCL2 also helps the inflammatory cells to migrate into the inflammatory site and the osteoclasts to differentiate.23 In this study, CXCL2 was upregulated 16.3-fold by the orthodontic force and 10.0-fold in the experimental group at 1 week after replantation, which decreased to the level in the control group at 3 weeks. This might indicate that bone remodeling reactions, including osteoclastogenesis, were promoted through the migration of inflammatory cells and osteoclast differentiation induced by CXCL2 during the 2 weeks after replantation. Previous studies have also shown that chemokine-driven cell migration is supposed to be involved in the remodeling of PDL tissue,24 and CXCL2 was highly expressed during orthodontic movement.25 Thus, our results were consistent with those described in previous reports.
In this study, CCL4 was upregulated at 1 week, followed by downregulation at 2, 3, and 4 weeks. CCL4 in a mouse model recruited bone marrow–derived monocytes to promote physiological revascularization.26 The active blood vessel formation seen in the experimental group in this study implied that CCL4 plays a role in revascularization during the healing process of the PDL tissue. In the control group, a histologically inflammatory reaction was sustained longer and the healing process was delayed. CCL7 is a small cytokine known to induce monocyte and macrophage migration27 and to regulate a wide variety of inflammatory cells.28 CCL7 was upregulated late (at 3 weeks) after replantation, whereas other chemokines were upregulated at 1 week. This might mean that CCL7 plays a role later in the process of tissue healing.
MMPs are enzymes involved in extracellular cleavage and activation of other MMPs and are responsible for different stages of degradation of the extracellular matrix.8 In particular, MMP3 has the ability to activate other MMPs, such as MMP8 and MMP9.29 The mRNA expression of MMP3 in the experimental group was upregulated 8.0-fold when compared with that in the control group at 1 week after replantation, and these levels remained upregulated for 2 weeks. Therefore, we can assume that MMP3 might be involved in the regeneration of PDL tissue during the healing process after transplantation in a time-dependent manner.
OPG as a decoy receptor for receptor activator of nuclear factor-κB (RANK) is produced by osteoblastic, hematopoietic, and immune cells and has been reported to suppress the activation of matrix osteoclasts by prohibiting binding between RANK and RANK ligand. RUNX2 has been confirmed as an osteoblast-specific transcription factor,30 and it plays an important regulatory role in the differentiation of mesenchymal stem cells into osteoblasts.31 The combined action of OPG and RUNX2 inhibits bone resorption and regulates the process of bone formation and bone resorption.32 The mRNA expression of OPG and RUNX2 was upregulated according to the same pattern in this study.
The results of this study showed that the application of orthodontic force before replantation caused the PDL tissue to proliferate and affected further healing of the replanted teeth. Even though a replantation model was used in this study owing to the difficulty in simulating the autotransplantation procedure in rats, it was assumed that the same healing process might occur after autotransplantation. Thus, the application of orthodontic force on the donor teeth before autotransplantation may result in a better healing process with less possibility of root resorption and ankylosis. Since this study mainly concerned time-related histological changes and mRNA expression levels using a replantation model and the possibility of ankyloses was not examined, future studies will be needed to verify how a biophysical force such as orthodontic force may affect PDL cell behavior at the molecular and biochemical levels and possibility of ankylosis.
This study investigated the effects of pre-applied orthodontic force on the tissue healing status after replantation and the results exhibited that application of orthodontic force prior to replantation enhanced the proliferation and activities of PDL cells. As the healing process in autotransplantation is assumed to be identical to that in replantation, pre-application of orthodontic force to donor teeth may also be recommended in autotransplantation to achieve better success rates.
Figures and Tables
Figure 1
Designs of the pilot study (top row) and the main study (bottom rows).
Lt, Left maxillary first molar (experimental group); Rt, right maxillary first molar (control group); PCR, polymerase chain reaction; H&E, hematoxylin-eosin.

Figure 2
A, Tissues stained with hematoxylin and eosin. a: The thickness of the PDL space is relatively even, and the alveolar bone surface is smooth. b: PDL tissues on the tension side are tensed and widened, whereas PDL tissues on the pressure side are narrow and compressed. A black arrow indicates force direction. B, Immunofluorescence staining showing reactivity to proliferating cell nuclear antigen (PCNA), white arrow indicates force direction. a: Control group without force. Moderate immunoreactivity to PCNA can be seen in the nuclei of PDL cells in the control group, and the cells in the alveolar bone and pulp exhibit weak reactivity. b: Experimental group in which orthodontic force was applied to the maxillary first molar for 1 week. PDL tissue spaces in the experimental group are widened after application of the orthodontic force. Numerous proliferating PDL cells show strong PCNA immunoreactivity in their nuclei (red arrows), as compared with the control group (a). Note that immunoreactivity in the pulpal cells is similar to that in the control group.
AB, Alveolar bone; D, dentin; PDL, periodontal ligament.
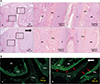
Figure 3
Histological changes in the periodontal tissue after replantation. All figures magnified from rectangular areas were acquired from the distal root of the first molar. One-, 2-, 3-, and 4-week control (A, C, E, G) and 1-, 2-, 3-, and 4-week experimental (B, D, F, H). The left and right black boxe are magnified in the middle and right columns, respectively, on each row. A, Roots were snugly fitted into the alveolar socket. Localized resorption (thick arrows) is seen at the apical and middle regions of the roots. The apical region of the distal root also showed cellular cementum regeneration. Of note, inflammatory cell infiltration (*) was seen near the blood vessels in the periodontal ligament (PDL), which shows deranged cell distribution. B, Many small blood vessels (arrows) are proliferating in the PDL space. However, few inflammatory features are seen. C, PDL space is widened owing to extensive root resorption (thick arrows). Moreover, localized infiltration of inflammatory cells (*) persists. D, PDL tissues appear normal. Cells are rearranged to form fiber groups, and blood vessels (arrows) have also reorganized on the side of the alveolar bone. E, Severe root resorption (thick arrows) is still ongoing. Infiltration of a number of inflammatory cells (*) persists. However, reorganization of the PDL cells and blood vessels (thin arrow) near the alveolar bone can be seen. F, The whole PDL tissue space appears even and regular. However, localized root resorption and cementum regeneration at the root apex are evident. G, Although root resorption (thick arrows) was still present, the PDL cells and blood vessels (thin arrows) became rearranged. Infiltration of inflammatory cells has subsided. H, Morphology of the roots appears normal, but PDL tissues appear to have matured normally.
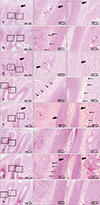
Figure 4
A, Expression of four genes was upregulated, as verified on microarray analysis, at 1 week after orthodontic tooth movement. B, Seven genes commonly related to periodontal ligament tissue remodeling (CXCL2, CCL4, CCL7, and MMP3), osteoclastogenesis (OPG), osteoblastogenesis (RUNX2), and cell proliferation (PCNA) were analyzed by means of real-time reverse transcription polymerase chain reaction. The expression levels of seven genes at 1, 2, 3, and 4 weeks after replantation are represented. Data (means ± standard deviation) were obtained from three independent experiments.
mRNA, Messenger RNA; CXCL2, macrophage inflammatory protein [MIP]-2α; CCL4, macrophage inflammatory protein [MIP]-1β; CCL7, monocyte chemotactic protein-3; MMP3, matrix metalloproteinase 3; OPG, osteoprotegerin; RUNX2, runt-related transcription factor 2; PCNA, proliferating cell nuclear antigen.
*p < 0.05; †p < 0.01; ‡p < 0.001 (by two-tailed Student's t-tests).

ACKNOWLEDGEMENTS
This study was carried out with the support of Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1D1A1B07051057 and NRF-2019R1A5A2027521).
Notes
References
1. Thilander B, Odman J, Lekholm U. Orthodontic aspects of the use of oral implants in adolescents: a 10-year follow-up study. Eur J Orthod. 2001; 23:715–731.

2. Schwartz O, Frederiksen K, Klausen B. Allotransplantation of human teeth. A retrospective study of 73 transplantations over a period of 28 years. Int J Oral Maxillofac Surg. 1987; 16:285–301.

3. Andreasen JO, Andersson L, Tsukiboshi M. Autotransplantation of teeth to the anterior region. In : Andreasen JO, Andreasen FM, Andreasen L, editors. Textbook and color atlas of traumatic injuries to the teeth. 4th ed. Copenhagen: Blackwell Munksgaard;2007. p. 740–760.
4. Andreasen JO, Paulsen HU, Yu Z, Schwartz O. A long-term study of 370 autotransplanted premolars. Part III. Periodontal healing subsequent to transplantation. Eur J Orthod. 1990; 12:25–37.

5. Andreasen JO. Periodontal healing after replantation and autotransplantation of incisors in monkeys. Int J Oral Surg. 1981; 10:54–61.

6. Huang L, Liu B, Cha JY, Yuan G, Kelly M, Singh G, et al. Mechanoresponsive properties of the periodontal ligament. J Dent Res. 2016; 95:467–475.

7. Kalajzic Z, Peluso EB, Utreja A, Dyment N, Nihara J, Xu M, et al. Effect of cyclical forces on the periodontal ligament and alveolar bone remodeling during orthodontic tooth movement. Angle Orthod. 2014; 84:297–303.

8. Han J, Xu X, Zhang B, Chen B, Hang W. Expression of ATF4 and RUNX2 in periodontal tissue of pressure side during orthodontic tooth movement in rat. Int J Clin Exp Med. 2015; 8:934–938.
9. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002; 30:e15.

10. Yang SY, Ko HM, Kang JH, Moon YH, Yoo HI, Jung NR, et al. Relaxin is up-regulated in the rat ovary by orthodontic tooth movement. Eur J Oral Sci. 2011; 119:115–120.

11. Andreasen JO. Analysis of pathogenesis and topography of replacement root resorption (ankylosis) after replantation of mature permanent incisors in monkeys. Swed Dent J. 1980; 4:231–240.
12. Schwartz O, Andreasen FM, Andreasen JO. Effects of temperature, storage time and media on periodontal and pulpal healing after replantation of incisors in monkeys. Dent Traumatol. 2002; 18:190–195.

13. Oswald RJ, Harrington GW, Van Hassel HJ. Replantation 1. The role of the socket. J Endod. 1980; 6:479–484.

14. Mine K, Kanno Z, Muramoto T, Soma K. Occlusal forces promote periodontal healing of transplanted teeth and prevent dentoalveolar ankylosis: an experimental study in rats. Angle Orthod. 2005; 75:637–644.
15. Andreasen JO. The effect of pulp extirpation or root canal treatment on periodontal healing after replantation of permanent incisors in monkeys. J Endod. 1981; 7:245–252.

16. Katayama A, Ota M, Sugito H, Shibukawa Y, Yamada S. Effect of proliferating tissue on transplanted teeth in dogs. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006; 101:e110–e118.

17. Ren Y, Maltha JC, Kuijpers-Jagtman AM. The rat as a model for orthodontic tooth movement--a critical review and a proposed solution. Eur J Orthod. 2004; 26:483–490.

18. Tomizuka R, Shimizu Y, Kanetaka H, Suzuki A, Urayama S, Kikuchi M, et al. Histological evaluation of the effects of initially light and gradually increasing force on orthodontic tooth movement. Angle Orthod. 2007; 77:410–416.

19. Suzaki Y, Matsumoto Y, Kanno Z, Soma K. Preapplication of orthodontic forces to the donor teeth affects periodontal healing of transplanted teeth. Angle Orthod. 2008; 78:495–501.

20. Bendre MS, Montague DC, Peery T, Akel NS, Gaddy D, Suva LJ. Interleukin-8 stimulation of osteoclastogenesis and bone resorption is a mechanism for the increased osteolysis of metastatic bone disease. Bone. 2003; 33:28–37.

21. Silva TA, Garlet GP, Fukada SY, Silva JS, Cunha FQ. Chemokines in oral inflammatory diseases: apical periodontitis and periodontal disease. J Dent Res. 2007; 86:306–319.

22. Maeda A, Bandow K, Kusuyama J, Kakimoto K, Ohnishi T, Miyawaki S, et al. Induction of CXCL2 and CCL2 by pressure force requires IL-1β-MyD88 axis in osteoblasts. Bone. 2015; 74:76–82.

23. Ha J, Lee Y, Kim HH. CXCL2 mediates lipopolysaccharide-induced osteoclastogenesis in RANKL-primed precursors. Cytokine. 2011; 55:48–55.

24. Masella RS, Meister M. Current concepts in the biology of orthodontic tooth movement. Am J Orthod Dentofacial Orthop. 2006; 129:458–468.

25. Madureira DF, Taddei Sde A, Abreu MH, Pretti H, Lages EM, da Silva TA. Kinetics of interleukin-6 and chemokine ligands 2 and 3 expression of periodontal tissues during orthodontic tooth movement. Am J Orthod Dentofacial Orthop. 2012; 142:494–500.

26. Ishikawa K, Yoshida S, Nakao S, Sassa Y, Asato R, Kohno R, et al. Bone marrow-derived monocyte lineage cells recruited by MIP-1β promote physiological revascularization in mouse model of oxygen-induced retinopathy. Lab Invest. 2012; 92:91–101.

27. Taub DD, Proost P, Murphy WJ, Anver M, Longo DL, van Damme J, et al. Monocyte chemotactic protein-1 (MCP-1), -2, and -3 are chemotactic for human T lymphocytes. J Clin Invest. 1995; 95:1370–1376.

28. Ben-Baruch A, Xu L, Young PR, Bengali K, Oppenheim JJ, Wang JM. Monocyte chemotactic protein-3 (MCP3) interacts with multiple leukocyte receptors. C-C CKR1, a receptor for macrophage inflammatory protein-1 alpha/Rantes, is also a functional receptor for MCP3. J Biol Chem. 1995; 270:22123–22128.

29. Beklen A, Tüter G, Sorsa T, Hanemaaijer R, Virtanen I, Tervahartiala T, et al. Gingival tissue and crevicular fluid co-operation in adult periodontitis. J Dent Res. 2006; 85:59–63.

30. Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997; 89:747–754.

SUPPLEMENTARY MATERIALS
Supplementary Table 1
Gene sequences examined on real-time reverse transcription polymerase chain reaction
Supplementary Table 2
Changes greater than 4-fold in genes of PDL tissue as a result of orthodontic force, as detected on microarray gene expression profiling
Supplementary Figure 1
The process of applying orthodontic force. A, A nickel-titanium (NiTi) coil spring was fixed to the first molar with ligature wire. B, The orthodontic force was measured with a tension gauge. C, D, The activated NiTi coil spring with 50 cN of force was fixed to the incisors with ligature wire and bonded with resin. E, The tip of the lower incisors was cut out to ensure stability of the NiTi coil spring. F, The application of orthodontic force was completed.
Supplementary Figure 2
The replantation procedure. A, B, Space between the first and second molars and tipping of the first molar were detected after successful application of orthodontic force. C–E, The first molar was gently extracted. F, After replantation of the first molar in the same socket, semi-rigid fixation was implemented.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download