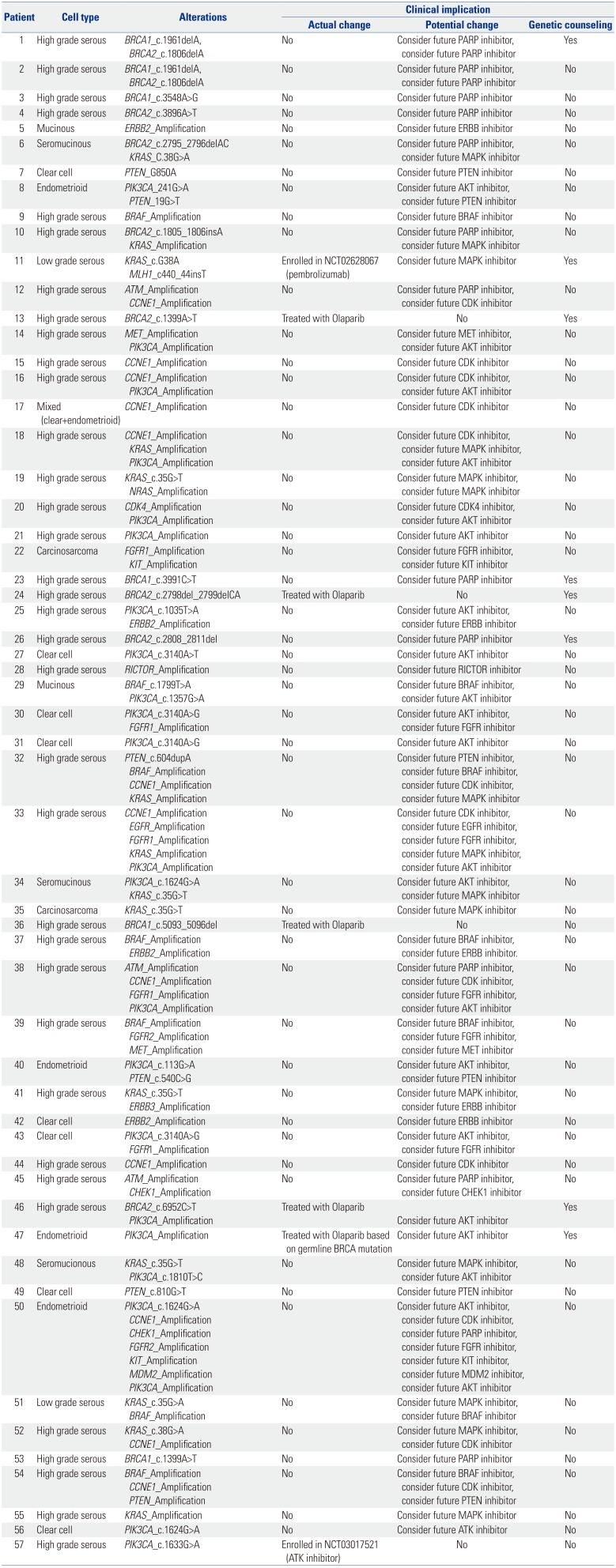
|
1 |
High grade serous |
BRCA1_c.1961delA, BRCA2_c.1806delA |
No |
Consider future PARP inhibitor, consider future PARP inhibitor |
Yes |
|
2 |
High grade serous |
BRCA1_c.1961delA, BRCA2_c.1806delA |
No |
Consider future PARP inhibitor, consider future PARP inhibitor |
No |
|
3 |
High grade serous |
BRCA1_c.3548A>G |
No |
Consider future PARP inhibitor |
No |
|
4 |
High grade serous |
BRCA2_c.3896A>T |
No |
Consider future PARP inhibitor |
No |
|
5 |
Mucinous |
ERBB2_Amplification |
No |
Consider future ERBB inhibitor |
No |
|
6 |
Seromucinous |
BRCA2_c.2795_2796delAC KRAS_C.38G>A |
No |
Consider future PARP inhibitor, consider future MAPK inhibitor |
No |
|
7 |
Clear cell |
PTEN_G850A |
No |
Consider future PTEN inhibitor |
No |
|
8 |
Endometrioid |
PIK3CA_241G>A PTEN_19G>T |
No |
Consider future AKT inhibitor, consider future PTEN inhibitor |
No |
|
9 |
High grade serous |
BRAF_Amplification |
No |
Consider future BRAF inhibitor |
No |
|
10 |
High grade serous |
BRCA2_c.1805_1806insA KRAS_Amplification |
No |
Consider future PARP inhibitor, consider future MAPK inhibitor |
No |
|
11 |
Low grade serous |
KRAS_c.G38A MLH1_c440_44insT |
Enrolled in NCT02628067 (pembrolizumab) |
Consider future MAPK inhibitor |
Yes |
|
12 |
High grade serous |
ATM_Amplification CCNE1_Amplification |
No |
Consider future PARP inhibitor, consider future CDK inhibitor |
No |
|
13 |
High grade serous |
BRCA2_c.1399A>T |
Treated with Olaparib |
No |
Yes |
|
14 |
High grade serous |
MET_Amplification PIK3CA_Amplification |
No |
Consider future MET inhibitor, consider future AKT inhibitor |
No |
|
15 |
High grade serous |
CCNE1_Amplification |
No |
Consider future CDK inhibitor |
No |
|
16 |
High grade serous |
CCNE1_Amplification PIK3CA_Amplification |
No |
Consider future CDK inhibitor, consider future AKT inhibitor |
No |
|
17 |
Mixed (clear+endometrioid) |
CCNE1_Amplification |
No |
Consider future CDK inhibitor |
No |
|
18 |
High grade serous |
CCNE1_Amplification KRAS_Amplification PIK3CA_Amplification |
No |
Consider future CDK inhibitor, consider future MAPK inhibitor, consider future AKT inhibitor |
No |
|
19 |
High grade serous |
KRAS_c.35G>T NRAS_Amplification |
No |
Consider future MAPK inhibitor, consider future MAPK inhibitor |
No |
|
20 |
High grade serous |
CDK4_Amplification PIK3CA_Amplification |
No |
Consider future CDK4 inhibitor, consider future AKT inhibitor |
No |
|
21 |
High grade serous |
PIK3CA_Amplification |
No |
Consider future AKT inhibitor |
No |
|
22 |
Carcinosarcoma |
FGFR1_Amplification KIT_Amplification |
No |
Consider future FGFR inhibitor, consider future KIT inhibitor |
No |
|
23 |
High grade serous |
BRCA1_c.3991C>T |
No |
Consider future PARP inhibitor |
Yes |
|
24 |
High grade serous |
BRCA2_c.2798del_2799delCA |
Treated with Olaparib |
No |
Yes |
|
25 |
High grade serous |
PIK3CA_c.1035T>A ERBB2_Amplification |
No |
Consider future AKT inhibitor, consider future ERBB inhibitor |
No |
|
26 |
High grade serous |
BRCA2_c.2808_2811del |
No |
Consider future PARP inhibitor |
Yes |
|
27 |
Clear cell |
PIK3CA_c.3140A>T |
No |
Consider future AKT inhibitor |
No |
|
28 |
High grade serous |
RICTOR_Amplification |
No |
Consider future RICTOR inhibitor |
No |
|
29 |
Mucinous |
BRAF_c.1799T>A PIK3CA_c.1357G>A |
No |
Consider future BRAF inhibitor, consider future AKT inhibitor |
No |
|
30 |
Clear cell |
PIK3CA_c.3140A>G FGFR1_Amplification |
No |
Consider future AKT inhibitor, consider future FGFR inhibitor |
No |
|
31 |
Clear cell |
PIK3CA_c.3140A>G |
No |
Consider future AKT inhibitor |
No |
|
32 |
High grade serous |
PTEN_c.604dupA BRAF_Amplification CCNE1_Amplification KRAS_Amplification |
No |
Consider future PTEN inhibitor, consider future BRAF inhibitor, consider future CDK inhibitor, consider future MAPK inhibitor |
No |
|
33 |
High grade serous |
CCNE1_Amplification EGFR_Amplification FGFR1_Amplification KRAS_Amplification PIK3CA_Amplification |
No |
Consider future CDK inhibitor, consider future EGFR inhibitor, consider future FGFR inhibitor, consider future MAPK inhibitor, consider future AKT inhibitor |
No |
|
34 |
Seromucinous |
PIK3CA_c.1624G>A KRAS_c.35G>T |
No |
Consider future AKT inhibitor, consider future MAPK inhibitor |
No |
|
35 |
Carcinosarcoma |
KRAS_c.35G>T |
No |
Consider future MAPK inhibitor |
No |
|
36 |
High grade serous |
BRCA1_c.5093_5096del |
Treated with Olaparib |
No |
No |
|
37 |
High grade serous |
BRAF_Amplification ERBB2_Amplification |
No |
Consider future BRAF inhibitor, consider future ERBB inhibitor. |
No |
|
38 |
High grade serous |
ATM_Amplification CCNE1_Amplification FGFR1_Amplification PIK3CA_Amplification |
No |
Consider future PARP inhibitor, consider future CDK inhibitor, consider future FGFR inhibitor, consider future AKT inhibitor |
No |
|
39 |
High grade serous |
BRAF_Amplification FGFR2_Amplification MET_Amplification |
No |
Consider future BRAF inhibitor, consider future FGFR inhibitor, consider future MET inhibitor |
No |
|
40 |
Endometrioid |
PIK3CA_c.113G>A PTEN_c.540C>G |
No |
Consider future AKT inhibitor, consider future PTEN inhibitor |
No |
|
41 |
High grade serous |
KRAS_c.35G>T ERBB3_Amplification |
No |
Consider future MAPK inhibitor, consider future ERBB inhibitor |
No |
|
42 |
Clear cell |
ERBB2_Amplification |
No |
Consider future ERBB inhibitor |
No |
|
43 |
Clear cell |
PIK3CA_c.3140A>G FGFR1_Amplification |
No |
Consider future AKT inhibitor, consider future FGFR inhibitor |
No |
|
44 |
High grade serous |
CCNE1_Amplification |
No |
Consider future CDK inhibitor |
No |
|
45 |
High grade serous |
ATM_Amplification CHEK1_Amplification |
No |
Consider future PARP inhibitor, consider future CHEK1 inhibitor |
No |
|
46 |
High grade serous |
BRCA2_c.6952C>T PIK3CA_Amplification |
Treated with Olaparib |
Consider future AKT inhibitor |
Yes |
|
47 |
Endometrioid |
PIK3CA_Amplification |
Treated with Olaparib based on germline BRCA mutation |
Consider future AKT inhibitor |
Yes |
|
48 |
Seromucionous |
KRAS_c.35G>T PIK3CA_c.1810T>C |
No |
Consider future MAPK inhibitor, consider future AKT inhibitor |
No |
|
49 |
Clear cell |
PTEN_c.810G>T |
No |
Consider future PTEN inhibitor |
No |
|
50 |
Endometrioid |
PIK3CA_c.1624G>A CCNE1_Amplification CHEK1_Amplification FGFR2_Amplification KIT_Amplification MDM2_Amplification PIK3CA_Amplification |
No |
Consider future AKT inhibitor, consider future CDK inhibitor, consider future PARP inhibitor, consider future FGFR inhibitor, consider future KIT inhibitor, consider future MDM2 inhibitor, consider future AKT inhibitor |
No |
|
51 |
Low grade serous |
KRAS_c.35G>A BRAF_Amplification |
No |
Consider future MAPK inhibitor, consider future BRAF inhibitor |
No |
|
52 |
High grade serous |
KRAS_c.38G>A CCNE1_Amplification |
No |
Consider future MAPK inhibitor, consider future CDK inhibitor |
No |
|
53 |
High grade serous |
BRCA1_c.1399A>T |
No |
Consider future PARP inhibitor |
No |
|
54 |
High grade serous |
BRAF_Amplification CCNE1_Amplification PTEN_Amplification |
No |
Consider future BRAF inhibitor, consider future CDK inhibitor, consider future PTEN inhibitor |
No |
|
55 |
High grade serous |
KRAS_Amplification |
No |
Consider future MAPK inhibitor |
No |
|
56 |
Clear cell |
PIK3CA_c.1624G>A |
No |
Consider future ATK inhibitor |
No |
|
57 |
High grade serous |
PIK3CA_c.1633G>A |
Enrolled in NCT03017521 (ATK inhibitor) |
No |
No |






 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download