Abstract
Purpose
Materials and Methods
Results
Figures and Tables
 | Fig. 1Cancer susceptibility candidate 2 (CASC2) significantly downregulated in hepatocellular carcinoma (HCC) tissues and cells. (A) The expression levels of CASC2 in 30 pairs of HCC tumor and matched neighboring non-tumor tissues (NTTs) were detected by real-time quantitative PCR (RT-qPCR). (B) Survival analysis in 30 HCC patients with high and low expression of CASC2. (C) CASC2 expression in human LO2 and five HCC cell lines (HepG2, Hep3B, QSG-7701, SMMC-7721 and Huh-7) was also analyzed by RT-qPCR. *p<0.05 compared to NTT; †p<0.05 compared to LO2 cells. |
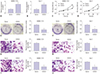 | Fig. 2Upregulation of cancer susceptibility candidate 2 (CASC2) inhibits cell viability, colony formation, migration, and invasion in hepatocellular carcinoma (HCC) cells in vitro. SMMC-7721 and Huh-7 cells were transfected with pcDNA3.1-CASC2 or pcDNA3.1 vector. The image magnification of transwell assay is ×100. The cells in colony formation assay and transwell assay were stained using crystal violet dye. (A and B) Real-time quantitative PCR was performed to analyze CASC2 expression in SMMC-7721 and Huh-7 cells after transfection. (C and D) MTT assay in SMMC-7721 and Huh-7 cells after transfection. (E and F) Colony formation assay in SMMC-7721 and Huh-7 cells after transfection. (G and H) Transwell assay detected migratory abilities of SMMC-7721 and Huh-7 cells after transfection. (I and J) Transwell assay detected the invasive abilities of SMMC-7721 and Huh-7 cells after transfection. *p<0.05 compared to cells transfected with pcDNA3.1 vector. OD, optical density. |
 | Fig. 3miR-183 is upregulated in hepatocellular carcinoma (HCC) tissues and cells. (A) Real-time quantitative PCR (RT-qPCR) was performed to analyze miR-183 expression in 30 pairs of HCC tumor and neighboring non-tumor tissues (NTTs). (B) RT-qPCR was conducted to analyze miR-183 expression in SMMC-7721 and Huh-7 cells. *p<0.05 compared to NTT; †p<0.05 compared to LO2 cells. |
 | Fig. 4miR-183 is directly targeted by cancer susceptibility candidate 2 (CASC2). (A) The sequence alignment of miR-183 with the 3′UTR of CASC2 is shown. (B and C) Luciferase assays in SMMC-7721 and Huh-7 cells transfected with wt CASC2 or mut CASC2 luciferase reporter plasmid, with miR-183 mimic or negative control (NC) mimic. (D and E) The expression of miR-183 in SMMC-7721 and Huh-7 cells transfected with pcDNA3.1-CASC2 or si-CASC2. (F) Correlation analysis between CASC2 and miR-183 in hepatocellular carcinoma (HCC) tissues, p<0.01. *p<0.05 compared to cells cotransfected with wt CASC2 and NC mimic; †p<0.05 compared to cells transfected with pcDNA3.1 vector or si-NC. |
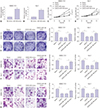 | Fig. 5Overexpression of miR-183 partially relieves cancer susceptibility candidate 2 (CASC2)-mediated inhibition on hepatocellular carcinoma (HCC) cell viability, colony formation, migration, and invasion. The image magnification of transwell assay is ×100. The cells in colony formation assay and transwell assay were stained using crystal violet dye. (A and B) Real-time quantitative PCR (RT-qPCR) assay for miR-183 expression in SMMC-7721 and Huh-7 cells transfected with negative control (NC) mimic or miR-183 mimic. (C-G) SMMC-7721 and Huh-7 cells were co-transfected with pcDNA3.1-CASC2 (CASC2) and NC mimic or miR-183 mimic. (C and D) MTT assay in SMMC-7721 and Huh-7 cells. (E) Colony formation assay in SMMC-7721 and Huh-7 cells. (F) Transwell assay detected the migratory abilities of SMMC-7721 and Huh-7 cells after transfection. (G) Transwell assay detected the invasive abilities of SMMC-7721 and Huh-7 cells after transfection. *p<0.05 compared to cells transfected with pcDNA3.1-vector or CASC2+NC mimic. OD, optical density. |
 | Fig. 6Cancer susceptibility candidate 2 (CASC2) regulates the Wnt/β-catenin pathway by downregulating miR-183. (A and B) Western blot assay for C-myc, cyclinD, surviving, and β-catenin expression in SMMC-7721 and Huh-7 cells co-transfected with pcDNA3.1-CASC2 or pcDNA3.1 vector with or without negative control (NC) mimic or miR-183 mimic. *p<0.05 compared to cells transfected with pcDNA3.1 vector; †p<0.05 compared to cells cotransfected with CASC2 and NC mimic. |
Notes
AUTHOR CONTRIBUTIONS
Conceptualization: Jian Sun and Lijun Liu.
Data curation: Jian Sun.
Formal analysis: Huilian Zou and Wei Yu.
Funding acquisition: Jian Sun.
Investigation: Wei Yu.
Methodology: Huilian Zou.
Project administration: Lijun Liu.
Resources: Huilian Zou.
Software: Wei Yu.
Supervision: Jian Sun.
Validation: Lijun Liu.
Visualization: Jian Sun.
Writing—original draft: Huilian Zou.
Writing—review & editing: Wei Yu.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download