Abstract
A variety of clonal cytogenetic abnormalities have been reported in aggressive natural killer (NK)-cell lymphoma and leukemia. Recent chromosomal microarray studies have shown both gain and loss of 1q and loss of 7p as recurrent abnormalities in aggressive NK-cell leukemia. Here, we report a case of aggressive NK-cell leukemia with complex chromosomal gains and losses, as confirmed by chromosomal microarray analysis. The patient showed an aggressive clinical course, which was complicated by hemophagocytic lymphohistiocytosis. Conventional cytogenetic analysis revealed trisomy 3 and 1q gain only. However, chromosomal microarray analysis detected an additional gain of 1q21.1-q24.2 and a loss of 1q24.2-q31.3. These abnormal lesions might play a role in the pathogenesis of aggressive NK-cell leukemia by inactivating tumor suppressor genes or by activating oncogenes. These results suggest that chromosomal microarray analysis may be used to provide further genetic information for patients with hematological malignancies, including aggressive NK-cell leukemia.
References
1. Nakashima Y, Tagawa H, Suzuki R, Karnan S, Karube K, Ohshima K, et al. Genomewide array-based comparative genomic hybridization of natural killer cell lymphoma/leukemia: different genomic alteration patterns of aggressive NK-cell leukemia and extranodal NK/T-cell lymphoma, nasal type. Genes Chromosomes Cancer. 2005; 44:247–55.

2. Chan JKC, Jaffe ES, Ko YH. Aggressive NK cell leukaemia. Swerdlow SH, Campo E, editors. WHO classifcation of tumours of haematopoietic and lymphoid tissues. Revised. 4th ed.Lyon: International Agency for Research on Cancer;2017. p. 353.
3. Siebert R. Mature B- and T-Cell neoplasms and Hodgkins lymphoma. Heim S, Mitelman F, editors. Cancer Cytogenetics. 3rd ed.Hoboken (N.J.): Wiley-Blackwell;2010. p. 336.
4. Siu LL, Wong KF, Chan JK, Kwong YL. Comparative genomic hybridization analysis of natural killer cell lymphoma/leukemia. Recognition of consistent patterns of genetic alterations. Am J Pathol. 1999; 155:1419–25.
5. Karube K, Nakagawa M, Tsuzuki S, Takeuchi I, Honma K, Nakashima Y, et al. Identifcation of FOXO3 and PRDM1 as tumor-suppressor gene candidates in NK-cell neoplasms by genomic and functional analyses. Blood. 2011; 118:3195–204.
6. Iqbal J, Kucuk C, Deleeuw RJ, Srivastava G, Tam W, Geng H, et al. Genomic analyses reveal global functional alterations that promote tumor growth and novel tumor suppressor genes in natural killer-cell malignancies. Leukemia. 2009; 23:1139–51.

7. Sako N, Dessirier V, Bagot M, Bensussan A, Schmitt C. HACE1, a potential tumor suppressor gene on 6q21, is not involved in extranodal natural killer/T-cell lymphoma pathophysiology. Am J Pathol. 2014; 184:2899–907.

8. Chang PW, Tsui SK, Liew C, Lee CC, Waye MM, Fung KP. Isolation, characterization, and chromosomal mapping of a novel cDNA clone encoding human selenium binding protein. J Cell Biochem. 1997; 64:217–24.

9. Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, et al. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003; 5:155–60.

10. Engel P, Eck MJ, Terhorst C. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat Rev Immunol. 2003; 3:813–21.

11. Kung C, Pingel JT, Heikinheimo M, Klemola T, Varkila K, Yoo LI, et al. Mutations in the tyrosine phosphatase CD45 gene in a child with severe combined immunodefciency disease. Nat Med. 2000; 6:343–5.
12. Simons C, Rash LD, Crawford J, Ma L, Cristofori-Armstrong B, Miller D, et al. Mutations in the voltage-gated potassium channel gene KCNH1 cause Temple-Baraitser syndrome and epilepsy. Nat Genet. 2015; 47:73–7.

13. Goueli BS, Powell MB, Finger EC, Pfeffer SR. TBC1D16 is a Rab4A GT-Pase activating protein that regulates receptor recycling and EGF receptor signaling. Proc Natl Acad Sci U S A. 2012; 109:15787–92.

14. Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004; 6:731–40.

15. Huang L, Liu D, Wang N, Ling S, Tang Y, Wu J, et al. Integrated genomic analysis identifes deregulated JAK/STAT-MYC-biosynthesis axis in aggressive NK-cell leukemia. Cell Res. 2018; 28:172–86.
Fig. 1.
In the peripheral blood smear, abnormal lymphoid cells resembling large granular lymphocytes are observed (Wright stain, ×1,000).
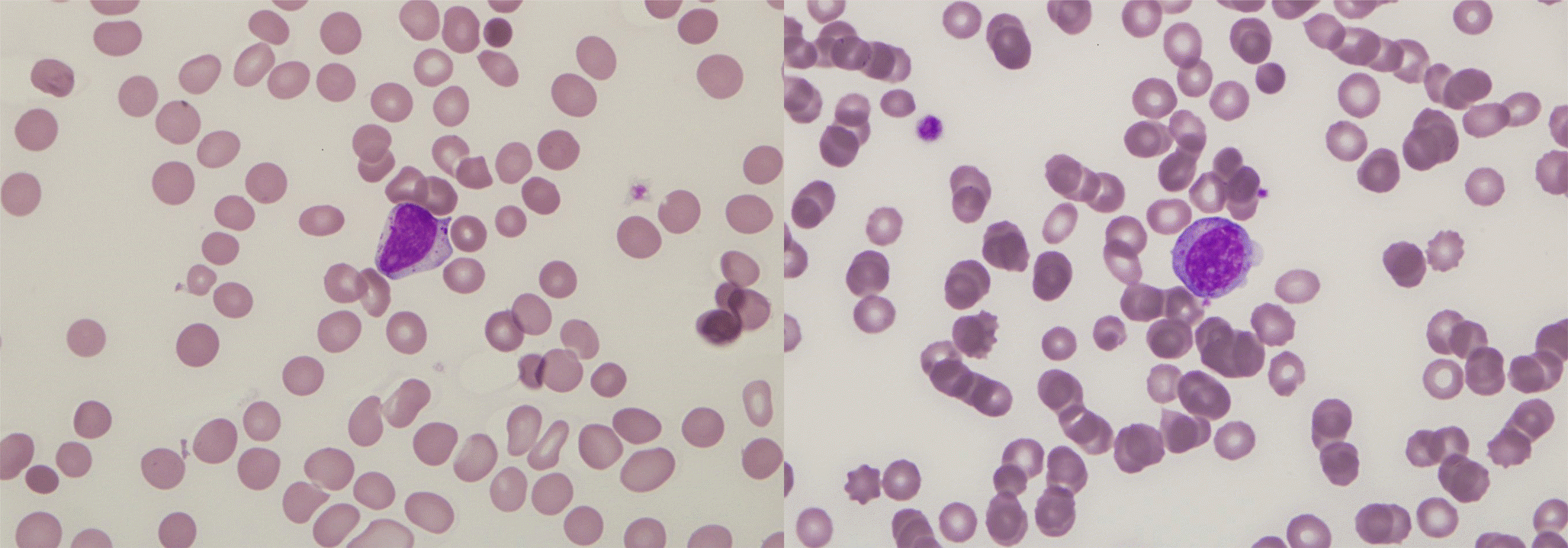
Fig. 2.
Immunophenotype analysis of leukemic cells using flow cytometry. Gated cells are 76.2% on side scatter vs. CD45 plot. Leukemic cells are positive for CD2, CD7 and CD56 and negative for surface CD3 (Cytomics FC500, Beckmann Coulter).
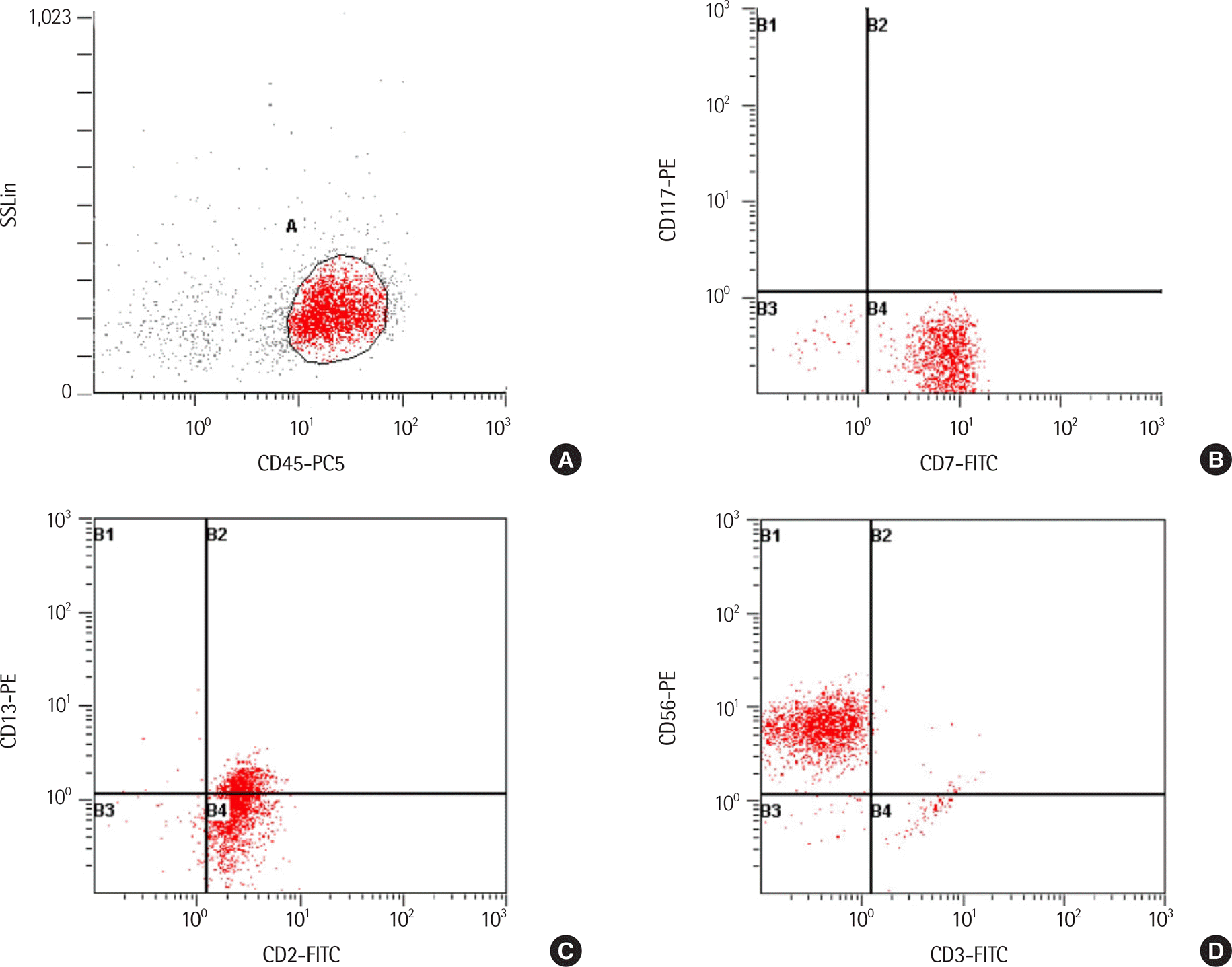
Fig. 5.
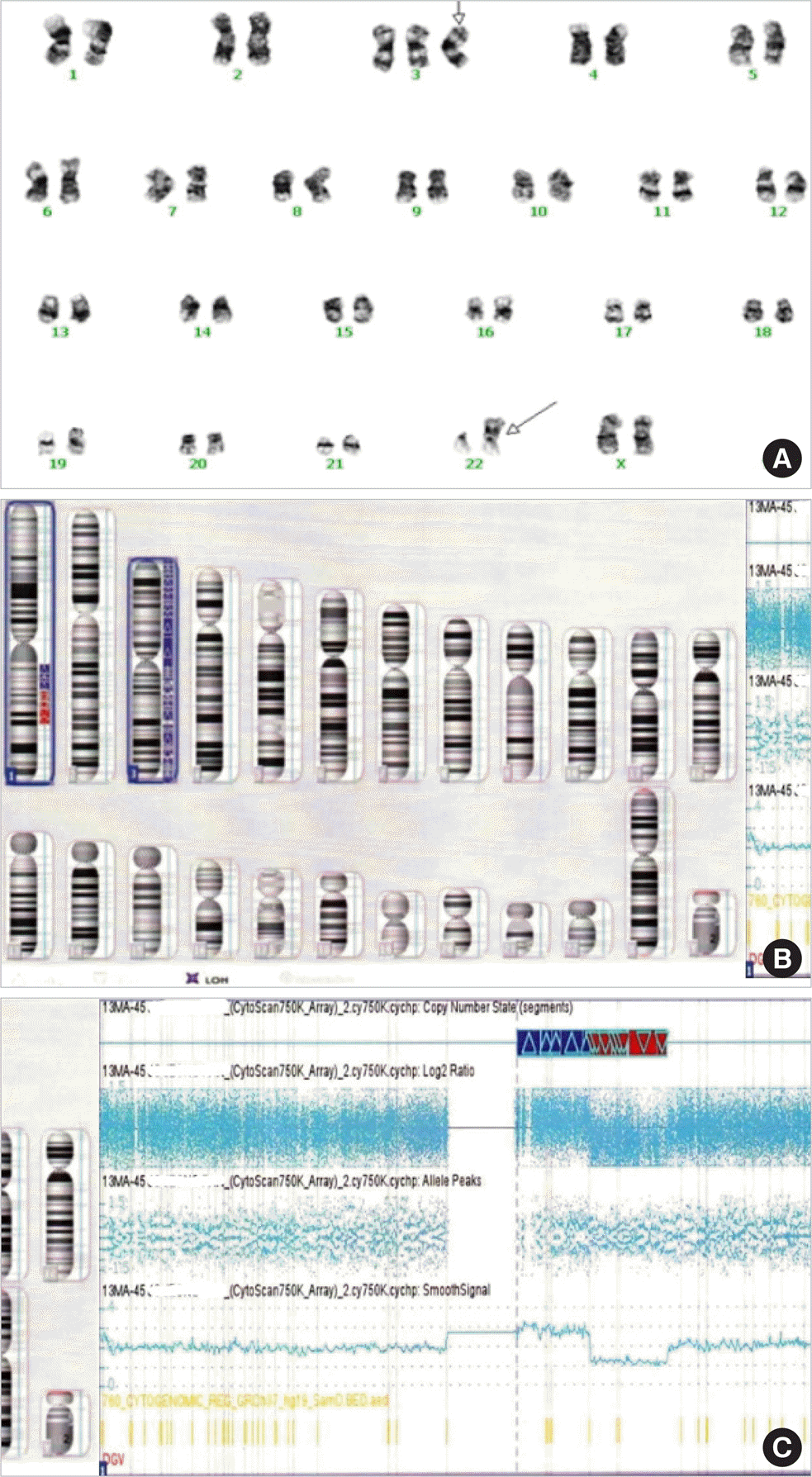
Karyotyping and chromosomal microarray analysis. (A) The karyotypes of peripheral blood cells at diagnosis by Giemsa banding. Arrows indicate abnormal chromosomes showing trisomy 3 and der(22)t(1;22)(q21;p11.2). (B) Idiogram of chromosomal microarray showing 1q gain and loss and trisomy 3. (C) Chromosomal microarray analysis of chromosome 1 showing 1q21.2-q24.2 gain of 24 Mb and 1q24.2-q31.3 loss of 26 Mb.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download