Abstract
Mycobacterium shimoidei is a nontuberculous mycobacterium (NTM), and is rarely reported as a pathogen causing the NTM pulmonary disease. We describe here the case of a 52-year-old male with symptoms such as chronic cough and a history of pulmonary tuberculosis. Radiologic studies revealed a cavitary lesion in the left upper lobe of his lung. Sputum culture was positive for NTM, which was later identified as M. shimoidei using 16S rRNA and hsp65 sequencing. The patient's symptoms, radiologic evidence, and positive culture results together substantiate that this is the first case of M. shimoidei pulmonary disease from Korea.
Nontuberculous mycobacterium (NTM) is a ubiquitously found organism, which is mainly responsible for the manifestation of the NTM lung disease. More than 180 species of NTM have been identified, and recent advances in molecular identification of bacteria along with the increasing number of cases of NTM infection have led to reports identifying other pathogenic subspecies of NTM [12]. Among the many reported species of NTM, only some are considered as respiratory pathogen, as environmental colonization of the respiratory tract is more frequently reported for some isolated strains of NTM [3]. Incidence and prevalence of the NTM disease shows global variation [45], and diagnosis of the NTM disease is more difficult when the isolated NTM does not have known clinical significance.
Mycobacterium shimoidei is a slow-growing NTM, which was first identified in 1975 with subsequent recognition as a species in 1983 [6]. M. shimoidei infection is very rare, and only 15 cases had been reported worldwide before a retrospective study involving 23 cases, including nine clinically significant cases, was performed in Australia in 2017 [7]. All three cases from Asia have been reported from Japan, including the first report of identification of M. shimoidei in 1975 [689]. Here, we report the first Korean case of pulmonary disease caused by M. shimoidei.
A 52-year-old male visited our hospital with chronic cough. The patient had a history of pulmonary tuberculosis and tuberculous pleurisy for 30 years; he also had a smoking history of 30 pack years. Two and a half years before his visit to our hospital, the patient had been diagnosed with M. avium lung disease and received antibiotic treatment with azithromycin, rifampin, and ethambutol for 13 months with conversion of sputum culture from positive to negative. At the time of diagnosis of the M. avium lung disease, NTM was isolated from two liquid culture samples. One was identified as M. avium and the other as unidentified Mycobacterium species, both using the line probe assay (LPA) for the rpoB region (REBA Myco-ID kit; YD diagnostics, Yongin, Korea).
At the time of the patient's visit to our hospital, high-resolution chest computed tomography (HRCT) revealed the development of a cavitary lesion in the left upper lobe of the lung (Fig. 1). Sputum cultures in liquid and solid media were performed using mycobacterial growth indicator tubes (MGIT 960 system; Becton Dickinson, Sparks, MD, USA) and 3% Ogawa agar (Shinyang, Seoul, Korea), and NTM were isolated from four liquid culture samples. For identification of the species of the causative Mycobacterium, LPA was performed for the internal transcribed spacer (ITS) region (AdvanSure Mycobacteria GenoBlot Assay; LG Chem, Seoul, Korea), revealing an unidentified Mycobacterium species [10]. For definitive identification of the species, 16S rRNA and 65 kDa heat shock protein genes (hsp65) from this isolate were sequenced using primer sets as previously described [10]. Direct sequencing was performed using the BigDye Terminator Cycle Sequencing Kit 3.1 (Applied Biosystems, Foster City, CA, USA) and the ABI Prism 3730 genetic analyzer (Applied Biosystems). After searching the GenBank database using the Basic Local Alignment Search Tool algorithm, we found that the 16S rDNA and hsp65 sequences of all four isolates exhibited 100% (518/518 bp) and 100% (401/401 bp) match, respectively, to those of M. shimoidei (GenBank accession no. AF547965.1 and AF547874.1). The next closest match was M. kyorinense at 97.5% (505/518 bp) and 95.5% (383/401 bp), respectively. A phylogenetic tree with a bootstrapping value of 1000 was reconstructed based on 16S rDNA and hsp65 sequences using the MEGA7 software (https://www.megasoftware.net) (Fig. 2). Drug susceptibility testing (DST) was performed at the Korean Institute of Tuberculosis using the broth microdilution method as described by the Clinical and Laboratory Standards Institute [11], the minimum inhibitory concentrations (µg/mL) of the antimicrobial agents used for DST are shown in Table 1.
After the final diagnosis of NTM lung disease caused by M. shimoidei, antibiotic treatment was initiated using azithromycin, ethambutol, and rifampicin. The patient completed 15 months of treatment along with clinico-radiological improvement and negative conversion of sputum culture.
The NTM pulmonary disease manifests as two major clinical phenotypes, the fibrocavitary and nodular bronchiectactic forms. The patient referred to in this report had acquired the typical fibrocavitary form of the NTM lung disease, which usually presents as pulmonary cavitation with a history of lung disease, including tuberculosis or malignancy. There are insufficient clinical data for M. shimoidei infection based on which methods of therapy might be applied; however, there have been reports on the successful use of ethambutol, rifabutin, and clarithromycin and resistance to rifampicin [121314].
M. shimoidei has often been misidentified as Mycobacterium terrae or Mycobacterium malmoense, using biochemical tests and HPLC methods, and was later identified correctly using 16S rRNA/DNA sequencing [131415]. LPAs can be used to detect 23–46 NTM species, and there are also several case reports wherein LPA has been used for identification of M. shimoidei [71016]. NTM identification is currently performed using either LPA or multigene sequence typing, and multigene sequence typing targeting 16S rDNA, 16S–23S ITS, rpoB, and hsp65 is known to be the most superior test available to distinguish between species [17]. The GenoType Mycobacterium CM/AS assay (GenoType assay, Hain Life-science, Nehren, Germany) is the only method available in Korea that can be used to identify M. shimoidei.
Interestingly, while M. shimoidei has been reported in 23 cases in a retrospective case series in Australia [7], and three times in Japan [8], none has been reported in Korea. While this can be attributed to the epidemiologic difference between the NTM species among different countries or regions [18], another possible explanation is that M. shimoidei is being identified incorrectly worldwide using HPLC, biochemical tests, and LPA.
Diagnosis of the NTM lung disease requires clinical/radiological and microbiological evidence, and M. shimoidei infection without concurrent clinical evidence does not fulfill the criteria for diagnosis of the NTM lung disease. Among the cases of M. shimoidei infection reported, majority of the cases were either considered clinically insignificant or reported without repeated culture positivity [7], samples from our patient however, were repeatedly found to be positive for sputum culture with aggravating symptoms and cavitary disease as found using HRCT.
As recurrent NTM infection is common, and recurrences are frequently associated with multiple species or genotypes, rarely reported NTM species will be more significant especially if patients have a history of the NTM disease [19]. DST for miscellaneous slowly growing NTM species, including M. shimoidei is recommended to be performed and interpreted as of rifampin-resistant Mycobacterium kansasii, owing to its rare occurrence [20]. The therapeutic strategy for these slowly growing species is generally similar to that used for the M. avium complex, and there are case reports of favorable outcomes with several different treatment regimens [16].
This is the first case report of M. shimoidei pulmonary disease from Korea and the second from Asia (the first being from Japan). Further investigation of the pathogenicity, drug susceptibility, and therapeutic strategy of M. shimoidei is required as it is a rare path-ogen.
Figures and Tables
Fig. 1
Mycobacterium shimoidei pulmonary disease in a 52-year-old male patient. Chest high-resolution computed tomography shows a thick-walled cavity in the left upper lobe of the lung. Note the associated emphysema in the left upper lobe, which suggests the sequelae of previous pulmonary tuberculosis.
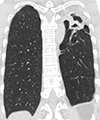
Fig. 2
Neighbor-joining phylogenetic tree showing the relationships of Mycobacterium shimoidei and other Mycobacterium species (A, 16S rDNA; B, hsp65 gene). Reference sequences are from species type strains; GenBank accession numbers are given in parentheses.

References
1. Parte AC. LPSN--list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014; 42:D613–D616.

2. Haworth CS, Banks J, Capstick T, Fisher AJ, Gorsuch T, Laurenson IF, et al. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). Thorax. 2017; 72:Suppl 2. ii1–ii64.

3. Aksamit TR, Philley JV, Griffith DE. Nontuberculous mycobacterial (NTM) lung disease: the top ten essentials. Respir Med. 2014; 108:417–425.

4. Ko RE, Moon SM, Ahn S, Jhun BW, Jeon K, Kwon OJ, et al. Changing epidemiology of nontuberculous mycobacterial lung diseases in a tertiary referral hospital in Korea between 2001 and 2015. J Korean Med Sci. 2018; 33:e65.

5. Stout JE, Koh WJ, Yew WW. Update on pulmonary disease due to non-tuberculous mycobacteria. Int J Infect Dis. 2016; 45:123–134.

6. Tsukamura M, Shimoide H, Shaefer WB. A possible new pathogen of group iii Mycobacteria. J Gen Microbiol. 1975; 88:377–380.

7. Baird TM, Carter R, Eather G, Thomson R. Mycobacterium shimoidei, a rare pulmonary pathogen, Queensland, Australia. Emerg Infect Dis. 2017; 23:1919–1922.
8. Saito H, Zayasu K, Shigeto E, Iwamoto T, Nakanaga K, Kodama A, et al. Two cases of lung infection due to Mycobacterium shimoidei, with special reference to bacteriological investigation. Kansenshogaku Zasshi. 2007; 81:12–19.

9. Takayama S, Tominaga S, Tsukada Y, Ohkochi M, Inase N. A case of pulmonary Mycobacterium shimoidei infection. Kekkaku. 2006; 81:537–541.
10. Yang M, Huh HJ, Kwon HJ, Kim JY, Song DJ, Koh WJ, et al. Comparative evaluation of the AdvanSure Mycobacteria GenoBlot assay and the GenoType Mycobacterium CM/AS assay for the identification of non-tuberculous mycobacteria. J Med Microbiol. 2016; 65:1422–1428.

11. Clinical and Laboratory Standards Institute. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Approved standard-2nd ed. Wayne, PA: Clinical and Laboratory Standards Institute;2011. CLSI document M24-A2.
12. Braganza Menezes DA, Dedicoat MJ, Robertson A. Mycobacterium shimoidei: an uncommon non-tuberculous infection in a UK patient. BMJ Case Rep. 2018; 2018:bcr-2017-221764.
13. Mayall B, Gurtler V, Irving L, Marzec A, Leslie D. Identification of Mycobacterium shimoidei by molecular techniques: case report and summary of the literature. Int J Tuberc Lung Dis. 1999; 3:169–173.
14. Heller R, Jaulhac B, Charles P, De Briel D, Vincent V, Bohner C, et al. Identification of Mycobacterium shimoidei in a tuberculosis-like cavity by 16S ribosomal DNA direct sequencing. Eur J Clin Microbiol Infect Dis. 1996; 15:172–175.

15. Sundman K, Chryssanthou E, Petrini B. Mycobacterium shimoidei, an easily misdiagnosed non-tuberculous pulmonary mycobacterium. Scand J Infect Dis. 2000; 32:450–451.
16. Popovic V, Arar D, Popovic DR, Barisic I, Tonkic M, Peric I, et al. Mycobacterium shimoidei-cavitary pulmonary disease with favorable outcome. Folia Microbiol (Praha). 2018; 63:249–252.

17. van Ingen J. Microbiological diagnosis of nontuberculous mycobacterial pulmonary disease. Clin Chest Med. 2015; 36:43–54.

18. Koh WJ, Kwon OJ, Jeon K, Kim TS, Lee KS, Park YK, et al. Clinical significance of nontuberculous mycobacteria isolated from respiratory specimens in Korea. Chest. 2006; 129:341–348.

19. Koh WJ, Moon SM, Kim SY, Woo MA, Kim S, Jhun BW, et al. Outcomes of Mycobacterium avium complex lung disease based on clinical phenotype. Eur Respir J. 2017; 50:1602503.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download