Abstract
A heart simulator, UT-Heart, is a finite element model of the human heart that can reproduce all the fundamental activities of the working heart, including propagation of excitation, contraction, and relaxation and generation of blood pressure and blood flow, based on the molecular aspects of the cardiac electrophysiology and excitation-contraction coupling. In this paper, we present a brief review of the practical use of UT-Heart. As an example, we focus on its application for predicting the effect of cardiac resynchronization therapy (CRT) and evaluating the proarrhythmic risk of drugs. Patient-specific, multiscale heart simulation successfully predicted the response to CRT by reproducing the complex pathophysiology of the heart. A proarrhythmic risk assessment system combining in vitro channel assays and in silico simulation of cardiac electrophysiology using UT-Heart successfully predicted druginduced arrhythmogenic risk. The assessment system was found to be reliable and efficient. We also developed a comprehensive hazard map on the various combinations of ion channel inhibitors. This in silico electrocardiogram database (now freely available at http://ut-heart.com/) can facilitate proarrhythmic risk assessment without the need to perform computationally expensive heart simulation. Based on these results, we conclude that the heart simulator, UT-Heart, could be a useful tool in clinical medicine and drug discovery.
With advancements in computational science bolstered by high-performance computing technology, it is now possible to reproduce the human heartbeat in a realistic three-dimensional simulation heart model implemented with mathematical models of functional molecules. The development and advancement of various novel diagnostic modalities require that heart simulators are not just multiscale but also multiphysics, to accurately compare with and examine multifaceted clinical data. Thus, we have developed a three-dimensional, multiscale, multiphysics heart simulator (UT-Heart). This simulator integrates the molecular and cellular mechanisms of cardiac excitation and contraction to reproduce the cardiac electrophysiology at the organ level and hemodynamics based on a finite element method with parallel computation [1].
First, we developed a method for analyzing fluid-structure interaction problems based on the arbitrary Lagrangian-Eulerian method [2], preconditioned iterative linear solvers for problems with constraint conditions [3], and a nonlinear homogenization method with low computational cost [4] as the numerical technique to realize the multiscale, multiphysics heart simulator. In our work, within the field of basic physiology, we also developed a three-dimensional cardiomyocyte-based model with the reproduction of detailed subcellular structures, including metabolism [56], a rational formula for accurate analysis of cardiac excitation propagation to overcome self-contraction of the bidomain equation [7], an excitation-contraction coupling model with the cooperativity effect [89], and an algorithm that optimizes the ventricular fiber structure of the human heart [10]. Moreover, we also investigated how the sinus of Valsalva relieves abnormal stress on aortic valve leaflets [11], developed a new echo-tracking method with deformable regions of interest aimed at rational pattern matching [12], and clarified the mechanism by which line block is generated in a non-contact mapping system using a heart simulator [13].
For the analysis of electrical phenomena in the heart, we reproduced the normal 12-lead electrocardiogram (ECG) tracing by using the finite element method [14]. This involved using the original, parallel, multilevel technique for solving the bidomain equations on a human heart with the use of Purkinje fibers and a human torso model [15]. For the analysis of mechanical phenomena in the heart, we started with the simulation of blood flow, left ventricular wall motion, and the relationship between them by fluid-structure interaction [1617]. We also established a multiscale heart simulation method that integrates the microscopic stochastic sarcomere kinetics into the macroscopic heartbeat via the mesoscopic myocardial cell assembly using a supercomputer [1819]. Although assembling these components to construct a comprehensive whole heart model was expensive in terms of computation, we have successfully achieved this, as shown in Fig. 1. We have also been granted numerous patents for this work (e.g., [2021]).We now intend to put UT-Heart to practical use in clinical medicine and pharmacology. In this article, we will focus on its application in predicting the effect of cardiac resynchronization therapy (CRT) and assessing the proarrhythmic risk of drugs. First, we review patient-specific heart models with realistic three-dimensional morphology based on clinical data recorded before treating nine patients with heart failure and conduction delay using biventricular pacing. Each model was individualized to reproduce the surface electrocardiography, echocardiography, and hemodynamic measurements corresponding to each patient. Next, we reviewed a system for assessing proarrhythmic risk combining in vitro channel assays and in silico simulation of cardiac electrophysiology using the developed UT-Heart. Furthermore, we extend this approach and develop a comprehensive hazard map on the various combinations of ion channel inhibitors using the RIKEN's K computer.
Extensive clinical research and basic studies have suggested potential biomarkers associated with the therapeutic responses of individual patients treated with CRT. However, it was found that a significant proportion (~30%) of patients do not benefit from this invasive therapy. To overcome this problem, computer simulations of CRT have been widely used to study the mechanisms underlying the therapeutic effects of this approach [2223]. As described above, we have developed a heart simulator in which propagation of excitation, contraction and relaxation, development of pressure, and blood flow are reproduced based on molecular models of cardiac electrophysiology and excitation-contraction coupling. We also succeeded in performing patient specific simulation of body surface ECG before and after CRT [24]. By applying these technologies, we created an individualized simulation model of the heart to determine whether the effects of CRT could be predicted in a canine model of heart failure with a left bundle branch block [25]. We further extended this approach to test the predictive ability of our simulator as a tool for the patient-specific evaluation of pathophysiology in a retrospective study. For nine patients treated with CRT, we created patient-specific heart models of the failing heart with ventricular desynchrony based on the clinical data recorded before treatment, and performed CRT simulations according to the actual cardiac pacing protocol. The simulation results of the effects of CRT correlated well with the clinical parameters that reflected the therapeutic effect. Thus, we demonstrated the utility of patient-specific CRT simulation and its possible application in future clinical practice [26].
The outline of our multiscale, multiphysics heart simulator and the technique for its individualized application are shown in Fig. 2. Details of mathematical models and calculation methods are previously reported [26].
(1) Patient-specific three-dimensional finite element (FE) models of the ventricles and upper body (torso) were reconstructed using multi-slice computed tomography (multi-scale CT) or magnetic resonance imaging data.
(2) These ventricular models with realistic morphology and made using fiber structure were subdivided into FEs. The molecular models of the excitation-contraction coupling process and electrophysiological models representing endocardial, M, or epicardial cells were implemented for these elements.
(3) The standard 12-lead ECGs recorded from the patients before CRT were reproduced through simulation by identifying the earliest activation sites and timing of activation in an iterative manner.
(4) Each model was individualized to reproduce the measurements of hemodynamic monitoring for each patient. Simultaneously, the lumped parameter models of the systemic and pulmonary circulations and time-varying elastance models of the atria were connected to the FE heart model, and the parameters were adjusted accordingly for each patient.
(5) CRT simulation was performed for each heart model without making any changes to the heart or circulation parameters determined during baseline simulation. The positions of the pacing leads were estimated using biplane chest radiography or CT images, wherever available, and then adjusted to reproduce the 12-lead ECG for pacing.
From the simulation results, we calculated the width of the QRS, ejection fraction (EF) in the left ventricle, and maximum value of the time derivative of left ventricular pressure (dP/dtmax). Clinical outcome was evaluated according to the improvement in EF indicated on the ultrasound cardiogram.
Examples of the simulated and actual ECGs before and after CRT for the standard 12 leads are shown in Fig. 3A. The patient in Fig. 3 suffered from complete atrioventricular block and had already been implanted with a pacemaker in the right ventricle. The simulated ECGs agreed well with the actual recordings by reproducing a wide QRS pattern before treatment. The corresponding excitations and contractions in these ECGs are shown in Fig. 3B. As seen in the figure, normal excitation propagated only from the right ventricle before treatment (Fig. 3B, top row). Presence of biventricular pacemaker excited both the ventricles from the basal side (Fig. 3B, bottom row). The electromechanical simulation coupled with the systemic and pulmonary circulations allowed the evaluation of the hemodynamic effects of CRT. As a ventricular aneurysm caused due to the thinning of the myocardium can be corrected by biventricular pacing, CRT increased the EF with a concomitant rise in the end-systolic pressure, indicating improved contractility (end-systolic elastance) as shown in Fig. 3C. This improvement was further substantiated by an increase in dP/dtmax (Fig. 3D).
In the nine patients studied, CRT increased the EF by 8.7 ± 5.7% (range −19% to 41%), according to the clinical records. Simulated improvements in EF by CRT (ΔEFsim) weakly correlated with the clinically observed improvements in EF (Fig. 4A). The simulated improvements were also much smaller than those seen clinically. However, there was a tight correlation of the clinically observed improvement in EF and the simulated increase in dP/dtmax (ΔdP/dtmax sim) (Fig. 4B). These findings suggest that dP/dtmax (r = 0.94) is a better predictor of CRT responses than EF (r = 0.59). Although the simulation effectively reproduced the clinical changes in a QRS duration, the clinical and simulated changes in the QRS duration (ΔQRSsim and ΔQRSclin) did not correlate with the clinical improvement in EF (Fig. 4C and D). This discrepancy between electrical and mechanical improvements in dyssynchrony is consistent with earlier clinical observations.
Multiscale, custom-made heart simulation can reproduce most of the pathophysiology of the failing heart and responses to CRT, thereby, providing mechanistic insight into the pathophysiology of heart failure due to ventricular conduction block. Although this is a retrospective study with a limited number of subjects, the predictive ability of this simulation technique warrants further examination with regard to future clinical applications.
Cardiac arrhythmias are a serious adverse effect of commonly used drugs. Among these, torsade de pointes (TdP) is a rare but potentially lethal type of ventricular arrhythmia often caused by inhibition of repolarizing potassium current of cardiac myocytes. The historical incidence of drug-related sudden cardiac deaths led to the establishment of regulatory requirements for cardiotoxicity testing, which include the measurement of repolarizing potassium current through the human ether-a-go-go-related gene (hERG) potassium current, in vivo QT assay in an animal model, and the examination of QT interval in healthy volunteers (a thorough QT study). However, these tests are costly and can sometimes lead to false-positive results, therefore, to overcome these issues, regulatory agencies have announced the future implementation of a new paradigm incorporating computational integration of a multiple ionic current assay into a cardiomyocyte model [272829]. Although the use of a cellular model is expected to greatly facilitate the screening process, accurate prediction of arrhythmia due to complex interactions among different types of cells in the heart tissues requires organ-level simulation. We proposed a system for assessing risk of proarrhythmia by combining in vitro channel assays using automated patch clamp technique and an in silico simulation of cardiac electrophysiology using UT-Heart (Fig. 5) [30]. The inhibitory effects of drugs are analyzed for the six ion channels (rapid delayed rectifier potassium current, sodium current, L-type calcium current, slow delayed rectifier potassium current, inward-rectifier potassium current, and transient outward potassium current), and the effects of drugs under various plasma concentrations are reproduced by changing the parameter values of 22 million cells implemented in the heart model. Our system succeeded in accurately predicting the arrhythmogenicity of 12 drugs (quinidine, D,l-sotalol, amiodarone, E-4031, cisapride, astemizole, terfenadine, bepridil, moxifloxacin, verapamil, dofetilide, and ranolazine) known to increase the risk of TdP, through the in vitro measurement of blocking actions on multiple ionic currents by reproducing a realistic human 12-lead ECG in silico. This heart simulator predicted proarrhythmic risk more accurately than the QT interval and hERG test. While all high-risk drugs can cause TdP at relatively low plasma concentrations, arrhythmia was not observed with safe drugs, even at high concentrations. Therefore, no false positive results were identified in our analysis. Various in silico methods have been proposed for accurate assessment of the risk of TdP induction by drugs.
This system can be applied to any drug candidate, but the requirement of a high-performance computer is an obstacle to its widespread use. To overcome this problem, we present a comprehensive hazard map of drug-induced arrhythmia based on an exhaustive in silico ECG database of drug effects, developed using RIKEN's K computer with performance of over 10 petaflops (SPARC64 VIIIfx, 705,024 cores; Fujitsu Ltd., Kawasaki, Japan [31]) [32]. We varied the extent of blocking of 5 ionic currents, which are known to affect arrhythmogenicity, incrementally by 10%, including that of rapid delayed rectifier potassium current (IKr: 0% to 100%), fast (INa: 0% to 40%) and late (INa,L: for this current, only 0%, 25%, and 50% blocks were tested) components of the sodium current, L-type calcium current (ICa,L: 0% to 40%), and slow delayed rectifier potassium current (IKs: 0% to 100%). Parallel computing on the K computer allowed us to obtain 9,075 patterns of electrophysiological simulations in 72,600,000 core hours. For every combination of current block, we simulated the electrophysiological activity of the heart and the associated 12-lead ECG to create a five-dimensional map of arrhythmia risk, in which the coordinates of the map represented the extent of block of each current. As we could not find a method to show a five-dimensional map in a single panel, we visualized the results in a five-dimensional presentation containing three-dimensional subspaces, where regions of arrhythmia were superimposed with the QT interval distribution. As the concentration-block relationship of each drug was traceable along a trajectory in these spaces, we were able to evaluate ECG changes and arrhythmogenicity (Fig. 6). This in silico ECG database facilitates the assessment of ECG characteristics and proarrhythmic risk without the need to computationally perform expensive heart simulations. Using this database, we can analyze the effect of each ion channel on the ECG characteristics (i.e., QRS duration and Tpeak-end, JT, and QT intervals) and the effect of this, in turn, on the risk of arrhythmias. Furthermore, because the safety margin of drugs at each concentration is assessed by the distance to the region of arrhythmia in these spaces, the database can provide not just the proarrhythmic risk, but also the margin of safety of these drugs. This database is now freely available at http://ut-heart.com/.
Drug-induced cardiotoxicity is not the only arrhythmogenic risk factor, heart's pumping function is also an important parameter to consider in cardiotoxicity testing. With UT-Heart, we can analyze the effect of drugs on pumping function. Fig. 7 demonstrates the pumping function during an arrhythmia provocation test. We applied an extra stimulus to the right ventricular apex in the presence of 12 times the free effective therapeutic plasma concentration range of cisapride. This led to early afterdepolarization (EAD) being induced that evolved to sustained ventricular arrhythmia. The area of EAD increased followed by a gradual increase in the frequency of excitation. Finally, the excitation evolved to ventricular fibrillation through TdP. The recorded history of blood pressure and left ventricular volume did not match the phase with corresponding electrocardiogram for arrhythmia (Fig. 7). Furthermore, the proarrhythmogenic risk of drugs sometimes conflicted with the negative cardiac inotropic effect. This can be exemplified with the fact that verapamil was found to be safe for arrhythmia in our simulation, as recognized clinically, despite significant QT prolongation on IKr inhibition [30]. However, on simultaneous inhibition of the L-type calcium current, verapamil caused a reduction in Ca transient in a dose-dependent manner. Such reduction in Ca transients, at the cellular level, results in impairment of ventricular pumping function, as shown by the small pressure-volume loops in Fig. 8. Verapamil is, thus, considered safe for arrhythmia, but it shows a significant negative effect on inotropic agents.
We present a brief review of a method to predict the effect of CRT using a patient-specific simulator and a proarrhythmic risk assessment system combined with in vitro channel assays using automated patch clamp technique and an in silico simulation of cardiac electrophysiology. The patient-specific, multiscale heart simulation successfully predicted the response to CRT by reproducing the complex pathophysiology of the heart. With further verification, this technique may serve to be a useful tool in clinical decision-making. The database of cardiotoxicity under multiple ion channel block is freely available at http://ut-heart.com/; thus, it cannot just be used to assess the risk of drug candidates at any stage of R&D, but can also serve as a tool to design safe drugs without resorting to animal or clinical studies. Therefore, this database may constitute a high-throughput and cost-saving approach in the field of toxicology.
This 21st century approach based on heart simulation can revolutionize the process of drug discovery and decision-making in clinical medicine. This work is a breakthrough in computational biology, introducing a novel application of high-performance computation.
Figures and Tables
 | Fig. 1Multiscale, multiphysics heart simulation.The heart model at the top shows the propagation of excitation signals and the model in the middle shows the blood flow in the heart chamber. The corresponding electrocardiogram (ECG) and pressure history are shown at the bottom. Corresponding videos are available at http://ut-heart.com. LV, left ventricle.
|
 | Fig. 2Custom-made cardiac resynchronization therapy (CRT) simulation.Patient-specific multiscale models of the heart and torso were created according to clinical data that were obtained before CRT (development step). Biventricular pacing was performed in this model, and the calculated biomarkers were validated and assessed by comparing them with clinical data obtained after CRT (validation and assessment step). CT, computed tomography; MRI, magnetic resonance imaging; ECG, electrocardiogram; UCG, ultrasound cardiogram; SR: sarcoplasmic reticulum; LV, left ventricle; EF, ejection fraction.
|
 | Fig. 3Simulated effects of cardiac resynchronization therapy (CRT).(A) Standard 12-lead electrocardiogram (ECG) before (left) and after (right) CRT. In each panel, ECGs are compared between simulation (in silico, left column) and actual recordings (in vivo, right column). (B) Time-lapse images of the propagation of excitation and contraction before (top row) and after (bottom row) CRT. Numbers at the bottom indicate the time of the onset of excitation (in milliseconds). (C) Left ventricular (LV) pressure-volume relationships before (black line) and after (red line) CRT. (D) The time derivative of LV pressure before (black line) and after (red line) CRT.
|
 | Fig. 4Predictive ability of biomarkers (modified from Okada et al . J Mol Cell Cardiol 2017;108:17-23 [26]).(A) Clinically observed improvement in the ejection fraction (ΔEFclin) vs . simulated improvement in the ejection fraction (ΔEFsim). (B) Clinically observed improvement in the ejection fraction (ΔEFclin) vs. simulated improvement at maximum dP/dt (ΔdP/dtmax sim). (C) Clinically observed improvement in the ejection fraction (ΔEFclin) vs. simulated narrowing of QRS (ΔQRSsim). (D) Clinically observed improvement in the ejection fraction (ΔEFclin) vs. clinically observed narrowing of QRS (ΔQRSclin). r, correlation coefficient.
|
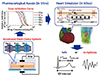 | Fig. 5A hybrid screening system for assessing drug-induced arrhythmogenic risk.Dose-inhibition curves of drugs were determined using the automated patch clamp system. On the basis of these data, conditions of the in silico heart model under specific drug concentrations were simulated by modulating the channel parameters in cell models to yield the 12-lead electrocardiogram.
|
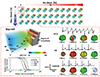 | Fig. 6Hazard map of drug-induced arrhythmias (modified from Okada et al. Br J Pharmacol 2018;175:3435-3452 [32]).The coordinate system of subspace consists of the extent of blocking of IKr, INa, and ICaL. In each subspace (in this case, IKs block = 0% and INaL block = 0%), the dose-dependent effect of the drug (bepridil) can be traced as a trajectory. The trajectories were generated by functions, and the numbers on them represent the drug concentrations expressed as multiples of the free effective therapeutic plasma concentration. The region of arrhythmia is indicated as brown blocks. Electrocardiogram changes at various concentrations are shown at the bottom with the corresponding activation sequence in the heart.
|
 | Fig. 7Drug-induced fatal arrhythmia.An extra stimulus was applied to the right ventricular apex at the end of the T-wave under cisapride administration. Premature ventricular contraction (PVC) evolved to ventricular fibrillation (VF) through torsade de pointes (TdP). Corresponding videos are available at http://ut-heart.com. ECG, electrocardiogram.
|
ACKNOWLEDGEMENTS
This work was supported in part by MEXT as a ‘Priority Issue on Post-K-computer’ (Integrated Computational Life Science to Support Personalized and Preventive Medicine, Project ID: hp150260, hp160209, and hp170233) and RIKEN as ‘Strategic Programs for Innovative Research Field 1’ (Computational Life Science and Application in Drug Discovery and Medical Development, Project ID: hp150236). The authors thank Edanz (www.edanzediting.co.jp) for editing the English text of a draft of this manuscript. We would like to thank Editage (www.editage.co.kr) for English language editing.
Notes
References
1. Sugiura S, Washio T, Hatano A, Okada J, Watanabe H, Hisada T. Multi-scale simulations of cardiac electrophysiology and mechanics using the University of Tokyo heart simulator. Prog Biophys Mol Biol. 2012; 110:380–389.

2. Zhang Q, Hisada T. Analysis of fluid-structure interaction problems with structural buckling and large domain changes by ALE finite element method. Comput Methods Appl Mech Eng. 2001; 190:6341–6357.

3. Washio T, Hisada T, Watanabe H, Tezduyar TE. A robust preconditioner for fluid-structure interaction problems. Comput Methods Appl Mech Eng. 2005; 194:4027–4047.

4. Okada J, Washio T, Hisada T. Study of efficient homogenization algorithms for nonlinear problems. Comput Mech. 2010; 46:247–258.

5. Okada J, Sugiura S, Nishimura S, Hisada T. Three-dimensional simulation of calcium waves and contraction in cardiomyocytes using the finite element method. Am J Physiol Cell Physiol. 2005; 288:C510–C522.

6. Hatano A, Okada J, Washio T, Hisada T, Sugiura S. A three-dimensional simulation model of cardiomyocyte integrating excitationcontraction coupling and metabolism. Biophys J. 2011; 101:2601–2610.

7. Okada J, Sugiura S, Hisada T. Modeling for cardiac excitation propagation based on the Nernst-Planck equation and homogenization. Phys Rev E Stat Nonlin Soft Matter Phys. 2013; 87:062701.

8. Washio T, Okada J, Sugiura S, Hisada T. Approximation for cooperative interactions of a spatially-detailed cardiac sarcomere model. Cell Mol Bioeng. 2012; 5:113–126.

9. Washio T, Sugiura S, Kanada R, Okada J, Hisada T. Coupling Langevin dynamics with continuum mechanics: exposing the role of sarcomere stretch activation mechanisms to cardiac function. Front Physiol. 2018; 9:333.

10. Washio T, Yoneda K, Okada J, Kariya T, Sugiura S, Hisada T. Ventricular fiber optimization utilizing the branching structure. Int J Numer Method Biomed Eng. 2016; 32:e02753.

11. Katayama S, Umetani N, Sugiura S, Hisada T. The sinus of Valsalva relieves abnormal stress on aortic valve leaflets by facilitating smooth closure. J Thorac Cardiovasc Surg. 2008; 136:1528–1535.

12. Cui X, Washio T, Chono T, Baba H, Okada J, Sugiura S, Hisada T. Deformable regions of interest with multiple points for tissue tracking in echocardiography. Med Image Anal. 2017; 35:554–569.

13. Okada J, Washio T, Nakagawa M, Watanabe M, Kadooka Y, Kariya T, Yamashita H, Yamada Y, Momomura SI, Nagai R, Hisada T, Sugiura S. Absence of rapid propagation through the Purkinje network as a potential cause of line block in the human heart with left bundle branch block. Front Physiol. 2018; 9:56.

14. Okada J, Washio T, Maehara A, Momomura S, Sugiura S, Hisada T. Transmural and apicobasal gradients in repolarization contribute to T-wave genesis in human surface ECG. Am J Physiol Heart Circ Physiol. 2011; 301:H200–H208.

15. Washio T, Okada J, Hisada T. A parallel multilevel technique for solving the bidomain equation on a human heart with Purkinje fibers and a torso model. SIAM Rev. 2010; 52:717–743.

16. Watanabe H, Hisada T, Sugiura S, Okada J, Fukunari H. Computer simulation of blood flow, left ventricular wall motion and their interrelationship by fluid-structure interaction finite element method. JSME Int J. 2002; 45:1003–1012.

17. Watanabe H, Sugiura S, Kafuku H, Hisada T. Multiphysics simulation of left ventricular filling dynamics using fluid-structure interaction finite element method. Biophys J. 2004; 87:2074–2085.

18. Hosoi A, Washio T, Okada J, Kadooka Y, Nakajima K, Hisada T. A multi-scale heart simulation on massively parallel computers. Paper presented at: SC '10: Proceedings of the 2010 ACM/IEEE International Conference for High Performance Computing, Networking, Storage and Analysis. 2010 Nov 13–19; New Orleans, LA, USA. IEEE Computer Society;p. 1–11.
19. Washio T, Okada J, Takahashi A, Yoneda K, Kadooka Y, Sugiura S, Hisada T. Multiscale heart simulation with cooperative stochastic cross-bridge dynamics and cellular structures. Multiscale Model Simul. 2013; 11:965–999.

20.
T Hisada
H Kurokawa
N Oshida
M Yamamoto
T Washio
J Okada
H Watanabe
S Sugiura
. Japan Science and Technology Agency. Modeling device, program, computer-readable recording medium, and method of establishing correspondence. United States patent. US 8,554,491. 2013.
21.
T Washio
T Hisada
E Nunobiki
J Okada
S Sugiura
. System for estimating membrane stress on arbitrarily-shaped curvilinear surface based on current configuration data. United States patent. US 9,241,639. 2016.
22. Lee AWC, Costa CM, Strocchi M, Rinaldi CA, Niederer SA. Computational modeling for cardiac resynchronization therapy. J Cardiovasc Transl Res. 2018; 11:92–108.

23. Niederer SA, Lumens J, Trayanova NA. Computational models in cardiology. Nat Rev Cardiol. 2019; 16:100–111.

24. Okada J, Sasaki T, Washio T, Yamashita H, Kariya T, Imai Y, Nakagawa M, Kadooka Y, Nagai R, Hisada T, Sugiura S. Patient specific simulation of body surface ECG using the finite element method. Pacing Clin Electrophysiol. 2013; 36:309–321.

25. Panthee N, Okada J, Washio T, Mochizuki Y, Suzuki R, Koyama H, Ono M, Hisada T, Sugiura S. Tailor-made heart simulation predicts the effect of cardiac resynchronization therapy in a canine model of heart failure. Med Image Anal. 2016; 31:46–62.

26. Okada J, Washio T, Nakagawa M, Watanabe M, Kadooka Y, Kariya T, Yamashita H, Yamada Y, Momomura SI, Nagai R, Hisada T, Sugiura S. Multi-scale, tailor-made heart simulation can predict the effect of cardiac resynchronization therapy. J Mol Cell Cardiol. 2017; 108:17–23.

27. Chi KR. Regulatory watch: Speedy validation sought for new cardiotoxicity testing strategy. Nat Rev Drug Discov. 2013; 12:655.
29. Gintant G, Sager PT, Stockbridge N. Evolution of strategies to improve preclinical cardiac safety testing. Nat Rev Drug Discov. 2016; 15:457–471.

30. Okada J, Yoshinaga T, Kurokawa J, Washio T, Furukawa T, Sawada K, Sugiura S, Hisada T. Screening system for drug-induced arrhythmogenic risk combining a patch clamp and heart simulator. Sci Adv. 2015; 1:e1400142.

31. Yonezawa A, Watanabe T, Yokokawa M, Sato M, Hirao K. Advanced Institute for Computational Science (AICS): Japanese national high-performance computing research institute and its 10-petaflops supercomputer “K”. Paper presented at: Proceedings of 2011 International Conference for High Performance Computing, Networking, Storage and Analysis. 2011 Nov 12–18; Seatle, WA, USA.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download