Abstract
Background
Carbapenem-resistant K. pneumoniae 2297, isolated from a patient treated with tigecycline for pneumonia, developed tigecycline resistance, in contrast to carbapenem-resistant isolate 1215, which was collected four months prior to the 2297 isolate. Mechanisms underlying tigecycline resistance were elucidated for the clinical isolates.
Methods
The tigecycline minimum inhibitory concentration (MIC) was determined using the broth microdilution method, with or without phenylalanine-arginine β-naphthylamide (PABN), and whole-genome sequencing was carried out by single-molecule real-time sequencing. The expression levels of the genes acrA,
oqxA,
ramA,
rarA, and rpoB were determined by reverse-transcription quantitative PCR.
Results
Both isolates presented identical antibiograms, except for tigecycline, which showed an MIC of 0.5 mg/L in 1215 and 2 mg/L in 2297. The addition of PABN to tigecycline-resistant 2297 caused a four-fold decrease in the tigecycline MIC to 0.5 mg/L, although acrA expression (encoding the AcrAB efflux pump) was upregulated by 2.5 fold and ramA expression (encoding the pump activator RamA) was upregulated by 1.4 fold. We identified a 6,096-bp fragment insertion flanking direct TATAT repeats that disrupted the romA gene located upstream of ramA in the chromosome of K. pneumoniae 2297; the insertion led the ramA gene promoter replacement resulting in stronger activation of the gene.
Klebsiella pneumoniae, a member of the Enterobacteriaceae family, causes both hospital- and community-acquired clinical infections, such as bloodstream infections and pneumonia, and exhibits antimicrobial resistance [1]. Drug treatment options, including tigecycline, are severely limited because of global dissemination of carbapenemase-producing K. pneumoniae, which confers resistance to carbapenems, considered as a last-resort treatment for infections [2].
Tigecycline is a minocycline-derivative semisynthetic glycylcycline with extended-spectrum anti-gram-negative activity. Tigecycline has been introduced for the treatment of community-acquired infections caused by extended-spectrum β-lactamase-producing Enterobacteriaceae [3]. Tigecycline can evade tetracycline and minocycline resistance mediated by the production Tet-type efflux pumps and TetM-mediated ribosomal protection [3]. Tigecycline resistance is mediated by the overproduction of resistance-nodulation-cell division (RND) efflux pumps, which have been identified in gram-negative bacteria: MexXY in Pseudomonas aeruginosa [4], SdeXY in Serratia marcescens [5], AcrAB and OqxAB in Enterobacter spp. [6], AcrAB and AcrEF in Escherichia coli [7], and AcrAB and OqxAB in K. pneumoniae [8]. The production of the AcrAB RND efflux pump in K. pneumoniae is locally regulated by the TetR-family transcriptional repressor, AcrR, and globally regulated by the AraC family transcriptional activators RamA, MarA, SoxS, and Rob [9]; the global regulators are prioritized over the local repressor [9]. Overproduction of another RND efflux system, OqxAB, is also regulated by the local repressor OqxR and the global activators RarA, MarA, SoxS, and Rob by overriding local regulation [10].
Two K. pneumoniae isolates producing NDM-5 and OXA-181 that exhibit indistinguishable XbaI-macrorestriction band patterns by pulsed-field gel electrophoresis were isolated within a span of four months in April and in August 2014, respectively, from epidemiologically unrelated patients hospitalized in a tertiary-care hospital in Korea [11]. The second patient was prescribed tigecycline for pneumonia treatment based on the antimicrobial susceptibility testing results; however, the causative K. pneumoniae isolate acquired tigecycline resistance within a few days of treatment, and the patient eventually died. The use of tigecycline to treat carbapenem-resistant K. pneumoniae infection has been reported to lead to the development of tigecycline resistance in accordance with the clinical setting [12]. We report a further analysis of the case for in vivo development of tigecycline resistance in those clinical isolates, as well as a putative hot spot for genetic recombination events conferring tigecycline resistance, based on whole genome comparisons.
K. pneumoniae 1215 (formerly CC1409-1) and K. pneumoniae 2297 (formerly CC1410-1) [11] isolates were used for the present retrospective study. The bacterial species of both isolates was confirmed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry using the MALDI Biotyper (Bruker Daltonics, GmbH, Bremen, Germany) and 16S rDNA sequencing. This research did not involve human subjects and was exempted from approval by the Ethics Committee on Human Research of the Health Ministry in Korea; the study design was not reviewed by an Institutional Review Board.
MLST of the K. pneumoniae isolates was performed by PCR and sequencing of seven house-keeping genes, namely, gapA,
infB,
mdh,
pgi,
phoE,
rpoB, and tonB [13]. The sequences obtained for both DNA strands were compared with sequences in the MLST database (http://bigsdb.web.pasteur.fr/klebsiella).
The minimum inhibitory concentrations (MICs) of aztreonam, cefotaxime, ceftazidime, cefoxitin, gentamicin, amikacin, ciprofloxacin, imipenem, meropenem, colistin, and tigecycline were determined by the broth microdilution method using freshly prepared cation adjusted Mueller-Hinton broth (CAMHB; Becton Dickinson, Franklin Lakes, NJ, USA) following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [14]. In addition, 20 mg/L of efflux pump inhibitor phenylalanine-arginine β-naphthylamide (PABN; Sigma-Aldrich, St. Louis, MO, USA) was added to determine whether the overproduced efflux pump system affects the tigecycline MIC. Escherichia coli ATCC 25922 and/or Pseudomonas aeruginosa ATCC 27853 were used for quality control, and the EUCAST breakpoints were adopted for clinical categorization [15]. The molecular mechanisms underlying carbapenem resistance were confirmed by PCR and sequencing of the blaOXA-48-like, blaIMP, blaVIM, blaGES, blaKPC, and blaNDM genes [16].
Total RNA was extracted from bacterial cells in the exponential growth phase (at an optical density at 600 nm, ca. 0.8) using the RNeasy plus mini kit (Qiagen, Hilden, Germany). The mRNA levels of acrA,
oqxA,
ramA,
rarA, and rpoB were quantified using a LightCycler 480 instrument II (Roche Diagnostics, Basel, Switzerland) with the LightCycler RNA amplification kit and SYBR Green I (Roche Diagnostics). The following gene-specific primers were designed and used for amplification: forward 5′-CAGGCAGCTTAGCGCTAACA-3′ and reverse 5′-CCTGGATATCGCTACCTTCC-3′ for acrA, forward 5′-TCCAGCGATAATCAGGCGCT-3′ and reverse 5′-CAGCGTGGCTTTGAACTCTG-3′ for oqxA, forward 5′-GCATCAACCGCTGCGTATTG-3′ and reverse 5′-ACGCGGGTAAAGGTCTGTTG-3′ for ramA, forward 5′-GGCGCCATCATTCAGGATCT-3′ and reverse 5′-AGTCAAAGCCGAGGGCAATC-3′ for rarA, and forward 5′-CTTGGTACGACCGTTCACGT-3′ and reverse 5′-GCTGAAACTGAACCACCTGG-3′ for rpoB. Amplification conditions consisted of one cycle of 95℃ for 30 seconds, followed by 45 cycles of 95℃ for five seconds, 58℃ for 10 seconds, and 72℃ for 20 seconds. The transcript expression levels of acrA,
oqxA,
ramA, and rarA were normalized against that of rpoB. Each experiment was performed in duplicate independently.
The complete genomes of K. pneumoniae 1215 and 2297 were sequenced by single-molecule real-time (SMRT) sequencing using a PacBio RSII instrument (Pacific Biosciences, Menlo Park, CA, USA). Genomic DNA was extracted from the K. pneumoniae isolates using the Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA). SMRTbell template libraries were subsequently prepared, and adapter ligation was performed. PacBio SMRT sequencing reads were used for de novo genome assembly with the PacBio SMRT analysis software suite (version 2.3.0). The circularized chromosome and plasmids were then polished using Quiver. Coding sequences, including tRNAs and rRNAs, were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/books/NBK174280). Nucleic acid sequences were compared using the Basic Local Alignment Search (BLAST) tool (http://blast.ncbi.nlm.nih.gov) [17], and resistance determinants and plasmid incompatibility types were determined using ResFinder (https://cge.cbs.dtu.dk/services/ResFinder) [18] and PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder) [19], respectively.
The K. pneumoniae 1215 and 2297 isolates were both identified as sequence type (ST) 147 by MLST. Both isolates exhibited carbapenem resistance conferred by both the blaOXA-181 and blaNDM-5 carbapenemase genes, as confirmed by PCR and sequencing. The isolates presented identical resistance profiles, except for tigecycline (MIC=0.5 mg/L for 1215 and 2 mg/L for 2297; Table 1). The tigecycline MIC for the quality control strain was 0.063 mg/L. The K. pneumoniae 1215 isolate was considered susceptible to tigecycline, whereas the 2297 isolate was considered resistant. Further, the isolates were resistant to aztreonam, cefotaxime, ceftazidime, cefoxitin, gentamicin, amikacin, and ciprofloxacin and susceptible to colistin.
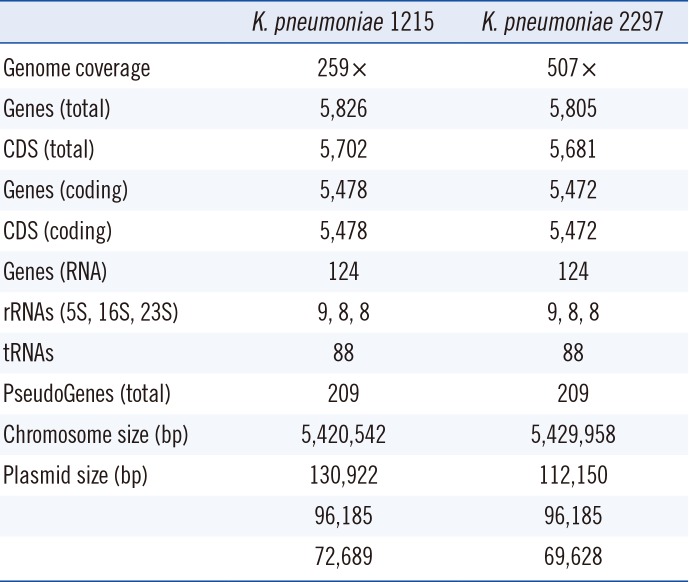
The whole-genome sequencing results showed that the full chromosomes of 1215 and 2297 (6,420,542 and 6,429,958 bp, respectively) shared 99% nucleic acid identity with 100% coverage for both sides (Table 2). A 9,416-bp additional segment found in the chromosome of 2297 comprised either insertion sequences or transposable DNA fragments. Two copies of blaOXA-181 were identified in each chromosome. The genomes of both 1215 and 2297 harbored the following three complete plasmids: identical 97,185 bp IncFII plasmids encoding NDM-5; 130,922 bp (1215) and 112,150 bp (2297) IncFII (pKPX1) plasmids, which were identical except for a 16,798-bp insert carrying five antimicrobial resistance genes, namely, aadA2, aacA4, and rmtF for aminoglycoside resistance, dfrA12 for trimethoprim resistance, and catA2 for chloramphenicol resistance; and nearly identical 72,689 bp (K. pneumoniae 1215) and 69,628 bp (K. pneumoniae 2297) non-incompatibility-typified cryptic plasmids.
The addition of the efflux pump inhibitor PABN led to a four-fold decrease in the tigecycline MIC from 2 to 0.5 mg/L for 2297. In contrast, no changes in MIC were observed for 1215. These results indicate that tigecycline resistance in 2297 is associated with overproduction of an efflux pump system.
We measured the expression levels of the acrA and oqrA genes, components of the operons encoding the AcrAB and OqrAB efflux systems, respectively, and of the activator-encoding ramA and rarA. We observed a 2.5-fold upregulation in acrA expression, consistent with a 1.4-fold upregulation of ramA expression in K. pneumoniae 2297 relative to that in 1215. No differences were observed in expression levels for the RND pump oqrA and the regulator rarA.
A 6,096-bp insertion flanking direct TATAT repeats was identified in the genome of K. pneumoniae 2297 that disrupted the RomA-coding sequences upstream of the ramA gene (Fig. 1). RomA is a repressor of the OmpF outer membrane protein [20], and the coding gene is located in the romA-ramA operon. The ramA gene has two transcriptional start sites, corresponding to the PI promoter upstream of the romA gene controlling romA-ramA operon transcription and the PII promoter located in the open reading frame (ORF) of romA, as evidenced by the presence of two size transcripts by northern blotting [21]. The original PII contains the −35 and −10 consensus sequences TTTATT and TATGGG, which are located 197 bp and 169 bp upstream of the transcriptional start site, respectively. Insertion of the 6,096-bp fragment in the romA ORF in K. pneumoniae 2297 introduced a new PII sequence harboring the −35 and −10 consensus sequences TTCCAT and GATAAT at the −133-bp and 104-bp regions upstream of the transcriptional start site.
The acrR gene, encoding a local repressor of AcrAB, was identical in both chromosomes. Furthermore, we could not identify amino acid alterations capable of conferring tigecycline resistance, such as in the tigecycline binding sites in the 16S rRNA [22] and ribosomal protein S10 [23]; global regulators RcsC, MarA, and SoxA; the ATP-binding cassette (ABC) transporter MacAB; or the outer membrane proteins OmpK35 and OmpK36.
We analyzed the mechanism of in vivo tigecycline resistance development, mostly through whole genome analysis, highlighting the need for cautious use of tigecycline for patients with infectious diseases.
Our findings clearly demonstrate that tigecycline resistance developed in vivo, and we identified the relevant genetic events. Subsequent development of tigecycline resistance in K. pneumoniae following tigecycline treatment in patients has been demonstrated in a cohort study involving 260 patients with carbapenem-resistant K. pneumoniae bacteriuria [24] and a case-control study on patients who tested positive for multi-drug resistant K. pneumoniae [25]. A significant relationship between tigecycline treatment and resistance development has been observed with an odds ratio of 6.13 (95% confidence interval [CI], 1.15–48.65) [24] and 6.00 (95% CI, 2.17–16.59) [25]. Tigecycline constitutes the final treatment option for pneumonia in patients infected by K. pneumoniae 2297 [11]. K. pneumoniae ST147 isolates are likely to persist in clinical settings by producing multiple carbapenemases and developing tigecycline resistance via genetic recombination during tigecycline treatment. Promoter replacement was likely responsible for the activation of the RND efflux pump, which in turn conferred tigecycline resistance in K. pneumoniae
in vivo.
Our study had several limitations. First, the approach was mostly observational. The carbapenem-resistant K. pneumoniae 2297 developed very strong resistance to tigecycline via the loss of romA. As colistin is the only useful selection marker in this species, a romA-complement plasmid could not be introduced; thus, the molecular microbiological proof was limited. Second, an epidemiological study of the tigecycline-resistant K. pneumoniae isolates was not conducted. The isolation of two carbapenemase producers with indistinguishable genomic profiles within four months warrants conducting an active surveillance study, especially as one of the isolates exhibited a broader spectrum of antimicrobial resistance. We investigated only the collected bacterial isolates, and the persistence of bacterial isolates in the hospital was not traced sufficiently and comprehensively.
In conclusion, our findings showed that in vivo development of tigecycline resistance was facilitated by the overproduction of the efflux pump. These results emphasize the need for careful selection of antimicrobial options for infectious diseases.
Notes
References
1. Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013; 11:297–298. PMID: 23458769.
2. Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015; 59:5873–5884. PMID: 26169401.
3. Livermore DM. Tigecycline: what is it, and where should it be used? J Antimicrob Chemother. 2005; 56:611–614. PMID: 16120626.
4. Dean CR, Visalli MA, Projan SJ, Sum PE, Bradford PA. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother. 2003; 47:972–978. PMID: 12604529.
5. Hornsey M, Ellington MJ, Doumith M, Hudson S, Livermore DM, Woodford N. Tigecycline resistance in Serratia marcescens associated with up-regulation of the SdeXY-HasF efflux system also active against ciprofloxacin and cefpirome. J Antimicrob Chemother. 2010; 65:479–482. PMID: 20051474.
6. Veleba M, De Majumdar S, Hornsey M, Woodford N, Schneiders T. Genetic characterization of tigecycline resistance in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Antimicrob Chemother. 2013; 68:1011–1018. PMID: 23349441.
7. Hirata T, Saito A, Nishino K, Tamura N, Yamaguchi A. Effects of efflux transporter genes on susceptibility of Escherichia coli to tigecycline (GAR-936). Antimicrob Agents Chemother. 2004; 48:2179–2184. PMID: 15155219.
8. Veleba M, Schneiders T. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2012; 56:4466–4467. PMID: 22644034.
9. Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2005; 49:1017–1022. PMID: 15728897.
10. De Majumdar S, Veleba M, Finn S, Fanning S, Schneiders T. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013; 57:1603–1609. PMID: 23318802.
11. Cho SY, Huh HJ, Baek JY, Chung NY, Ryu JG, Ki CS, et al. Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg Infect Dis. 2015; 21:1088–1089. PMID: 25988911.
12. Du X, He F, Shi Q, Zhao F, Xu J, Fu Y, et al. The Rapid Emergence of Tigecycline Resistance in blaKPC-2 Harboring Klebsiella pneumoniae, as Mediated in Vivo by Mutation in tetA During Tigecycline Treatment. Front Microbiol. 2018; 9:648. PMID: 29675006.
13. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005; 43:4178–4182. PMID: 16081970.
14. EUCAST. Antimicrobial susceptibility testing of bacteria. Updated on Jan 2017. http://www.eucast.org/ast_of_bacteria/.
15. EUCAST. Breakpoint tables for interpretation of MICs and zone diameters. version 7.1. Updated on Mar 2017. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf.
16. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011; 70:119–123. PMID: 21398074.
17. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. PMID: 9254694.
18. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012; 67:2640–2644. PMID: 22782487.
19. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014; 58:3895–3903. PMID: 24777092.
20. Komatsu T, Ohta M, Kido N, Arakawa Y, Ito H, Kato N. Increased resistance to multiple drugs by introduction of the Enterobacter cloacae romA gene into OmpF porin-deficient mutants of Escherichia coli K-12. Antimicrob Agents Chemother. 1991; 35:2155–2158. PMID: 1662028.
21. Rosenblum R, Khan E, Gonzalez G, Hasan R, Schneiders T. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int J Antimicrob Agents. 2011; 38:39–45. PMID: 21514798.
22. Bauer G, Berens C, Projan SJ, Hillen W. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA. J Antimicrob Chemother. 2004; 53:592–599. PMID: 14985271.
23. Fang L, Chen Q, Shi K, Li X, Shi Q, He F, et al. Step-Wise increase in tigecycline resistance in Klebsiella pneumoniae associated with mutations in ramR,
lon and rpsJ. PLoS One. 2016; 11:e0165019. PMID: 27764207.
24. van Duin D, Cober ED, Richter SS, Perez F, Cline M, Kaye KS, et al. Tigecycline therapy for carbapenem-resistant Klebsiella pneumoniae (CRKP) bacteriuria leads to tigecycline resistance. Clin Microbiol Infect. 2014; 20:O1117–O1120. PMID: 24931918.
25. Nigo M, Cevallos CS, Woods K, Flores VM, Francis G, Perlman DC, et al. Nested case-control study of the emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013; 57:5743–5746. PMID: 23979745.
Fig. 1
Schematic representation of the insertion of a 6,096 bp DNA fragment disrupting the romA gene in Klebsiella pneumoniae 2297. Gray boxes indicate identical sequences between K. pneumoniae 1215 and 2297. Gene names are indicated above or below the open arrows. The direct TATAT repeats are indicated above their location.
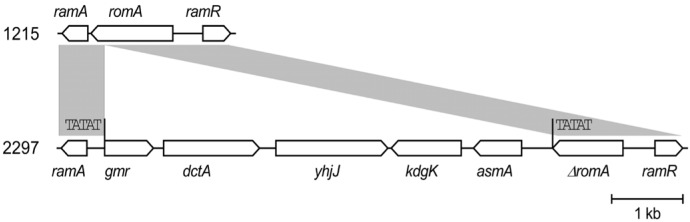
Table 1
Antimicrobial susceptibility of the Klebsiella pneumoniae ST147 isolates
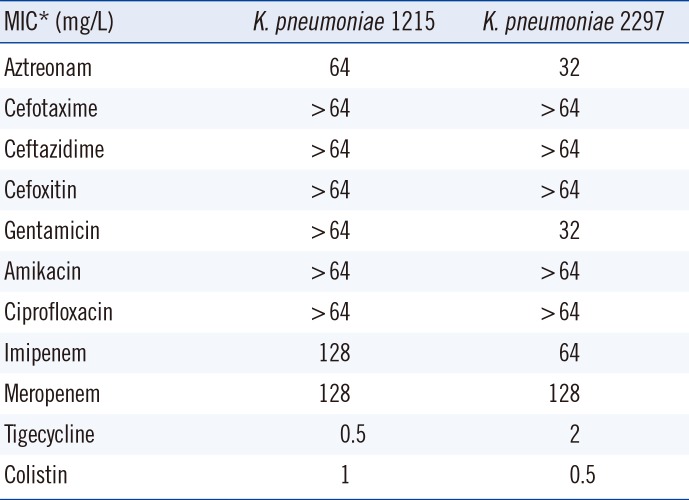
Table 2
Comparison between the genomes of K. pneumoniae 1215 and 2297




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download