This article has been
cited by other articles in ScienceCentral.
Abstract
Accurate detection of BCR-ABL fusion transcripts at and below molecular response (MR) 4 (0.01% International Scale [IS]) is required for disease monitoring in patients with chronic myeloid leukemia (CML). We evaluated the analytical performance of the QXDx BCR-ABL %IS (Bio-Rad, Hercules, CA, USA) droplet digital PCR (ddPCR) assay, which is the first commercially available ddPCR-based in vitro diagnostics product. In precision analysis, the %CV was 9.3% and 3.0%, with mean values of 0.031% IS and 9.4% IS, respectively. The assay was linear in the first order, ranging from 0.032% IS to 20% IS. The manufacturer-claimed limit of blank, limit of detection, and limit of quantification were verified successfully. There was a very strong correlation between the results of the QXDx BCR-ABL %IS ddPCR assay and the ipsogen BCR-ABL1 Mbcr IS-MMR (Qiagen, Hilden, Germany) real-time quantitative PCR assay (r=0.996). In conclusion, the QXDx BCR-ABL %IS ddPCR assay can provide reliable results for CML patients.
Go to :

Keywords: BCR-ABL, Droplet digital PCR, Performance, Evaluation, Chronic myeloid leukemia
Recent practice guidelines from the European LeukemiaNet and National Comprehensive Cancer Network for the management of patients with chronic myeloid leukemia (CML) call for the use of sensitive PCR assays, like real-time quantitative PCR (RQ-PCR) assays, for detecting
BCR-ABL fusion transcripts during treatment, monitoring minimal residual disease (MRD), and identifying patients at risk of relapse [
12]. It is recommended that patients are tested every three months and the results are reported in International Scale (% IS) units for standardized reporting of the molecular response (MR) [
3]. However, RQ-PCR assays are limited in terms of limit of detection (LOD) and limit of quantification (LOQ) [
4]. Undetectable
BCR-ABL fusion transcripts using RQ-PCR assays, especially at the LOD and LOQ, affect clinical decisions and may lead to inappropriate or premature cessation of treatment [
45]; adequate sensitivity levels should be achieved to detect MRD down to MR4 (0.01% IS) or MR4.5 (0.0032% IS) [
6].
Droplet digital PCR (ddPCR) assay can quantify the total copy number of targets present in a sample without standards [
7]. Although its underlying chemistry is similar to that of the RQ-PCR assay, the ddPCR assay has an additional step, which separates each sample into 20,000 nanoliter-sized droplets, in which the PCR occurs, improving assay precision and reproducibility [
789]. The QXDx BCR-ABL %IS (Bio-Rad, Hercules, CA, USA) is the first ddPCR-based
in vitro diagnostics (IVD) product with the US Food and Drug Administration clearance and European Conformity (CE) mark; however, its analytical performance has not been evaluated to date [
10]. We evaluated the precision, linearity, and detection capability (limit of blank [LOB], LOD, and LOQ) of the QXDx BCR-ABL %IS ddPCR assay. We also evaluated its correlation with the CE-IVD-marked ipsogen BCR-ABL1 Mbcr IS-MMR DX (Qiagen, Hilden, Germany) RQ-PCR assay, which has been designed according to the “Europe Against Cancer” studies and is compliant with the updated international recommendations [
111213].
This study was conducted between May and June 2019 at Konkuk University Medical Center (KUMC), Seoul, Korea, after obtaining exemption from approval by the Institutional Review Board of KUMC (KUH1200100). Venous whole blood (3 mL) was collected in K3 EDTA vacutainer (Greiner Bio-one, Kremsmünster, Austria), the BCR-ABL mRNA was extracted using the QIAamp RNA Blood Mini Kit (Qiagen, Valencia, CA, USA), and the mRNA samples were stored at −70℃ until use. Both the QXDx BCR-ABL %IS ddPCR assay and BCR-ABL Mbcr IS-MMR DX RQ-PCR assay were performed following the manufacturers' instructions. For the ddPCR assay, an Automated Droplet Generator (Bio-Rad), CFX96 thermal cycler (Bio-Rad), and QX200 Droplet Reader (Bio-Rad) were used; the raw data were analyzed and interpreted using the QuantaSoft software 1.7.4 (Bio-Rad).
Precision of the QXDx BCR-ABL %IS ddPCR assay was determined using a low positive control (MR3.0–5.0) and ~10% IS calibrator (MR1.0), which were included in the kit, according to the CLSI guidelines EP15-A3, using manufacturer-claimed within laboratory imprecision [
14]. We replicated the assay three times in a single run, on three separate days. The %CVs were 9.3% and 3.0% with mean values of 0.031% IS and 9.4% IS, respectively; the maximal %CV of 9.3% was used to define allowable errors in the linearity assessment [
14].
Linearity was determined at five levels (20, 10, 1.0, 0.1, and 0.032% IS) using a certified reference material (CRM) for BCR-ABL1, ERM-AD623f (European Commission's Joint Research Centre, EU); the CRM was diluted using CML-negative human blood samples, according to the CLSI guidelines EP06-A [
15]. Quantification using the QXDx BCR-ABL %IS ddPCR assay showed a linear shape in the first order (y=0.9981x+0.0305) in log scale, ranging from 0.032% IS to 20% IS. No outliers were detected by visual examination of the scatter plot. The observed data were within the allowable error (<9.3%) with a CV of 2.6%, 1.4%, 3.9%, 7.6%, and 4.8% at the levels of 20, 10, 1.0, 0.1, and 0.032% IS, respectively.
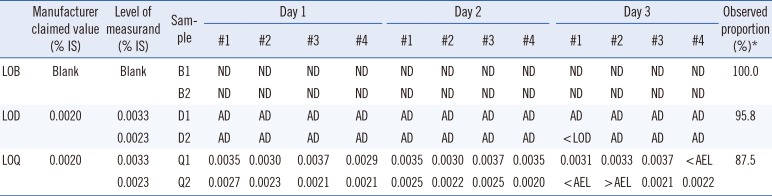
Detection capability was estimated according to the CLSI guidelines EP17-A2 [
16]. The total number of measurements was 24 each for LOB, LOD, and LOQ. To verify the LOB claim, two blank samples were measured with four replicates per sample on three separate days using one reagent lot; all 24 (100%) replicates showed the result “not detected.” To verify the LOD and LOQ claims (0.002% IS [MR4.7] for both), the lowest linearity material was diluted using CML-negative human blood samples to 0.0033% IS (MR4.5) and 0.0023% IS (MR4.6), respectively. Each sample was measured with four replicates on three separate days using one reagent lot. For LOD verification, 23 (95.8%) of the 24 replicates showed the result “analyte detected” and only one replicate was lower than the LOD (<0.002% IS). For LOQ verification, 21 (87.5%) of the 24 replicates were within the allowable error window (accuracy goal of ±15% total error at a level of 0.0028–0.0038% IS for Q1 and 0.0020–0.0027% IS for Q2;
Table 1). The observed LOD and LOQ proportions (95.8% and 87.5%) all exceeded the minimum percentage of 85% (95% confidence interval) with a sample size of 24 [
16], verifying the manufacturer's LOB, LOD, and LOQ claims.
Table 1
Verification of the detection capability claims of the QXDx BCR-ABL %IS ddPCR assay
|
Manufacturer claimed value (% IS) |
Level of measurand (% IS) |
Sample |
Day 1 |
Day 2 |
Day 3 |
Observed proportion (%)*
|
|
#1 |
#2 |
#3 |
#4 |
#1 |
#2 |
#3 |
#4 |
#1 |
#2 |
#3 |
#4 |
|
LOB |
Blank |
Blank |
B1 |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
100.0 |
|
B2 |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
ND |
|
LOD |
0.0020 |
0.0033 |
D1 |
AD |
AD |
AD |
AD |
AD |
AD |
AD |
AD |
AD |
AD |
AD |
AD |
95.8 |
|
0.0023 |
D2 |
AD |
AD |
AD |
AD |
AD |
AD |
AD |
AD |
< LOD |
AD |
AD |
AD |
|
LOQ |
0.0020 |
0.0033 |
Q1 |
0.0035 |
0.0030 |
0.0037 |
0.0029 |
0.0035 |
0.0030 |
0.0037 |
0.0035 |
0.0031 |
0.0033 |
0.0037 |
< AEL |
87.5 |
|
0.0023 |
Q2 |
0.0027 |
0.0023 |
0.0021 |
0.0021 |
0.0025 |
0.0022 |
0.0025 |
0.0020 |
< AEL |
> AEL |
0.0021 |
0.0022 |

The quantitative results of the QXDx BCR-ABL %IS ddPCR assay and BCR-ABL Mbcr IS-MMR DX RQ-PCR assay were compared using Passing-Bablok regression analysis according to the CLSI guidelines EP09-A3 [
17]. Using a total of 20 clinical samples (ranging from 0.002% IS [MR4.7] to 20% IS [MR0.7]), the results of the two assays demonstrated a very strong correlation (r=0.996;
Fig. 1).
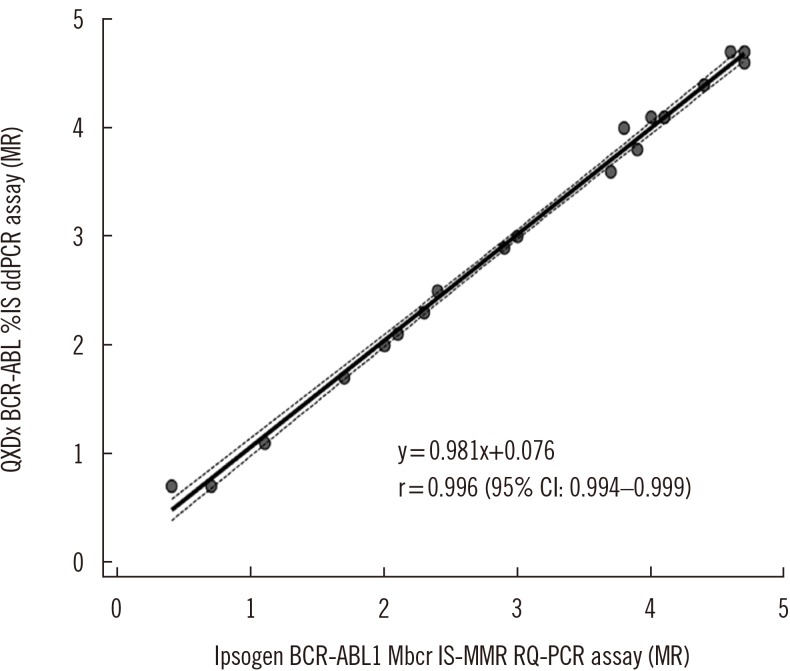 | Fig. 1
Passing-Bablok regression of the correlation between the QXDx BCR-ABL %IS ddPCR assay and the BCR-ABL Mbcr IS-MMR DX RQ-PCR assay (N=20). Regression line with a 95% CI is shown.
Abbreviations: ddPCR, droplet digital PCR; RQ-PCR, real-time quantitative PCR; MR, molecular response; CI, confidence interval.

|
One limitation of the QXDx BCR-ABL %IS ddPCR assay was that it was designed to detect only the e13a2 and e14a2 fusion transcripts, but not e1a2, e19a2, or other rare transcripts. Other potential limitations or disadvantages are its longer turnaround time due to the additional time required for droplet generation (60–70 min/plate) and droplet reading (120–140 min/plate) and the possibility of false positivity, although we observed no false positives [
718]. Owing to the limited number of available assay kits, LOD and LOQ were verified at the levels of 0.0023% IS (MR4.64) and 0.0033% IS (MR4.49), and we could not test levels <0.002% IS (MR4.7). Further studies are needed to verify the clinical and laboratory utility of the QXDx BCR-ABL %IS ddPCR assay.
In conclusion, this is the first study to evaluate the analytical performance of the novel QXDx BCR-ABL %IS ddPCR assay. With its acceptable analytical performance, the QXDx BCR-ABL %IS ddPCR assay can be a reliable and promising tool for MRD monitoring in CML patients.






 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download