Linezolid, the first approved oxazolidinone antibiotic by the US Food and Drug Administration for clinical use, is an important treatment option in cases of multidrug-resistant gram-positive bacteria, including methicillin-resistant
Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci [
1]. Linezolid inhibits protein synthesis by binding to the 50S subunit of the ribosome via the domain V region of the 23S ribosomal RNA (rRNA) [
23]. Mutations in domain V have been associated with linezolid resistance in MRSA (e.g., G2447T, T2500A, and G2576T;
Escherichia coli numbering system). The G2576T mutation (most frequently detected in clinical isolates) is associated with prolonged use of linezolid-combined antibiotic treatment [
12].
S. aureus contains five or six copies of the 23S rRNA gene, and the level of resistance appears to be associated with an increasing number of mutant copies [
24]. Recently, other molecular mechanisms of linezolid resistance in MRSA have been reported in clinical isolates, including mutations in ribosomal proteins L3, L4, and L22 encoded by the
rplC,
rplD, and
rplV genes, respectively [
35]. In addition, plasmid-mediated resistance, including the
optrA,
cfr (chloramphenicol-florfenicol resistance), and
cfr(B) genes, has been described as a resistance mechanism underlying horizontal transmission between different species [
678].
As various linezolid resistance mechanisms have been identified, we investigated the molecular characterization of linezolid-resistant MRSA for the first time in Korea. We determined the linezolid resistance and molecular characteristics of MRSA with elevated linezolid minimum inhibitory concentrations (MICs) as per the VITEK 2 system (bioMérieux, Marcy-l'Étoile, France), one of the most widely used automated antimicrobial susceptibility testing systems.
A total of 22,067 MRSA isolates were obtained from January 2014 to December 2018 at Samsung Medical Center, Seoul, Korea, and the antimicrobial susceptibility test results were retrospectively reviewed. Of these 22,067 MRSA isolates, only 110 (0.5%) were linezolid-resistant, with linezolid MICs of ≥8 µg/mL in VITEK 2. Of these, 27 isolates from 14 patients were stored in skim milk at −70℃ following routine susceptibility testing and were available for this study. These isolates were identified using VITEK 2 or VITEK MS (bioMérieux). All relevant clinical data were collected for analysis, including dose and length of linezolid treatment and other antimicrobial susceptibility test results. This study was approved by the Institutional Review Board (IRB) of Samsung Medical Center (IRB no. SMC 2016-05-048), and informed consent requirements were waived.
Automated antimicrobial susceptibility testing was performed using the VITEK 2 AST-P601 card (bioMérieux) according to the manufacturer's instructions. The MIC of the 27 linezolid-resistant clinical isolates was confirmed using the broth microdilution (BMD) test [
9]. The panel for the BMD test was prepared by lyophilizing linezolid in a commercial 96-well cell culture plate (SPL Life Sciences, Pocheon, Korea). The BMD test was performed using two-fold serial dilutions ranging from 0.5 to 128 µg/mL. Inoculation, incubation, and interpretation were based on the CLSI guidelines [
910].
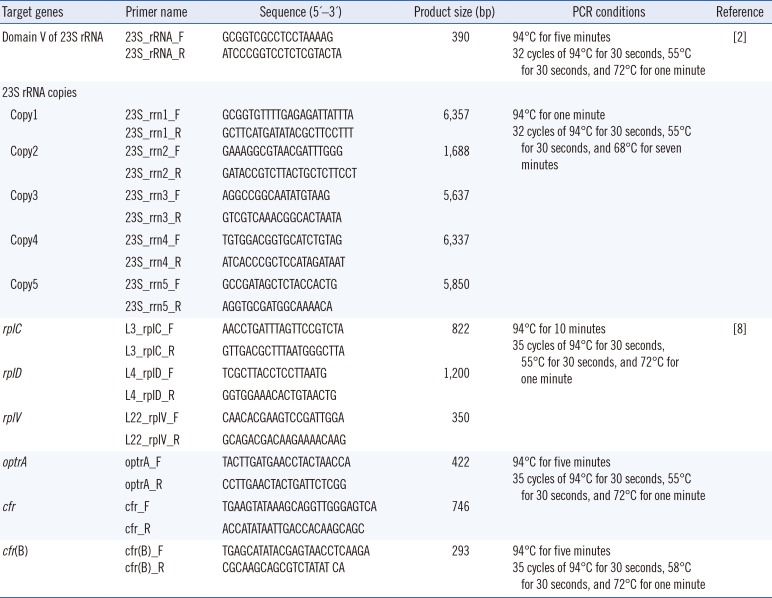
Genomic DNA was extracted from the linezolid-resistant clinical isolates using the Nextractor NX-48 nucleic acid extraction system (Genolution, Seoul, Korea). Mutations in domain V of the 23S rRNA gene and ribosomal proteins (L3, L4, and L22) were investigated using PCR. Sequencing with specific primers and amplification conditions are shown in
Table 1. The acquired 23S rRNA-encoding DNA sequences and amino acid sequences of L3, L4, and L22 were compared with the
S. aureus reference sequence (GenBank accession No. NR_076325.1). If a mutation was detected in domain V of the 23S rRNA gene, each of the five copies of the 23S rRNA gene was amplified using a high-fidelity PCR premix (AccuPower Premix; Bioneer, Daejeon, Korea) for long-range PCR. In addition, the presence of the
optrA,
cfr, and
cfr(B) genes in the linezolid-resistant clinical isolates was investigated using oligonucleotide primers, following Lee, et al. [
8].
Table 1
Primer sequences and PCR conditions used for the amplification and sequencing of 23S rRNA, rplC, rplD, rplV, optrA, cfr, cfr(B), and 23S rRNA copies in Staphylococcus aureus, as well as amplicon product size of the amplified regions
|
Target genes |
Primer name |
Sequence (5′–3′) |
Product size (bp) |
PCR conditions |
Reference |
|
Domain V of 23S rRNA |
23S_rRNA_F |
GCGGTCGCCTCCTAAAAG |
390 |
94℃ for five minutes |
[2] |
|
23S_rRNA_R |
ATCCCGGTCCTCTCGTACTA |
32 cycles of 94℃ for 30 seconds, 55℃ for 30 seconds, and 72℃ for one minute |
|
23S rRNA copies |
|
|
|
|
|
|
Copy1 |
23S_rrn1_F |
GCGGTGTTTTGAGAGATTATTTA |
6,357 |
94℃ for one minute |
|
23S_rrn1_R |
GCTTCATGATATACGCTTCCTTT |
32 cycles of 94℃ for 30 seconds, 55℃ for 30 seconds, and 68℃ for seven minutes |
|
Copy2 |
23S_rrn2_F |
GAAAGGCGTAACGATTTGGG |
1,688 |
|
|
23S_rrn2_R |
GATACCGTCTTACTGCTCTTCCT |
|
Copy3 |
23S_rrn3_F |
AGGCCGGCAATATGTAAG |
5,637 |
|
|
23S_rrn3_R |
GTCGTCAAACGGCACTAATA |
|
Copy4 |
23S_rrn4_F |
TGTGGACGGTGCATCTGTAG |
6,337 |
|
|
23S_rrn4_R |
ATCACCCGCTCCATAGATAAT |
|
Copy5 |
23S_rrn5_F |
GCCGATAGCTCTACCACTG |
5,850 |
|
|
23S_rrn5_R |
AGGTGCGATGGCAAAACA |
|
rplC
|
L3_rplC_F |
AACCTGATTTAGTTCCGTCTA |
822 |
94℃ for 10 minutes |
[8] |
|
L3_rplC_R |
GTTGACGCTTTAATGGGCTTA |
35 cycles of 94℃ for 30 seconds, 55℃ for 30 seconds, and 72℃ for one minute |
|
rplD
|
L4_rplD_F |
TCGCTTACCTCCTTAATG |
1,200 |
|
|
L4_rplD_R |
GGTGGAAACACTGTAACTG |
|
rplV
|
L22_rplV_F |
CAACACGAAGTCCGATTGGA |
350 |
|
|
L22_rplV_R |
GCAGACGACAAGAAAACAAG |
|
optrA
|
optrA_F |
TACTTGATGAACCTACTAACCA |
422 |
94℃ for five minutes |
|
|
optrA_R |
CCTTGAACTACTGATTCTCGG |
35 cycles of 94℃ for 30 seconds, 55℃ for 30 seconds, and 72℃ for one minute |
|
cfr
|
cfr_F |
TGAAGTATAAAGCAGGTTGGGAGTCA |
746 |
|
|
cfr_R |
ACCATATAATTGACCACAAGCAGC |
|
cfr(B) |
cfr(B)_F |
TGAGCATATACGAGTAACCTCAAGA |
293 |
94℃ for five minutes |
|
|
cfr(B)_R |
CGCAAGCAGCGTCTATAT CA |
35 cycles of 94℃ for 30 seconds, 58℃ for 30 seconds, and 72℃ for one minute |

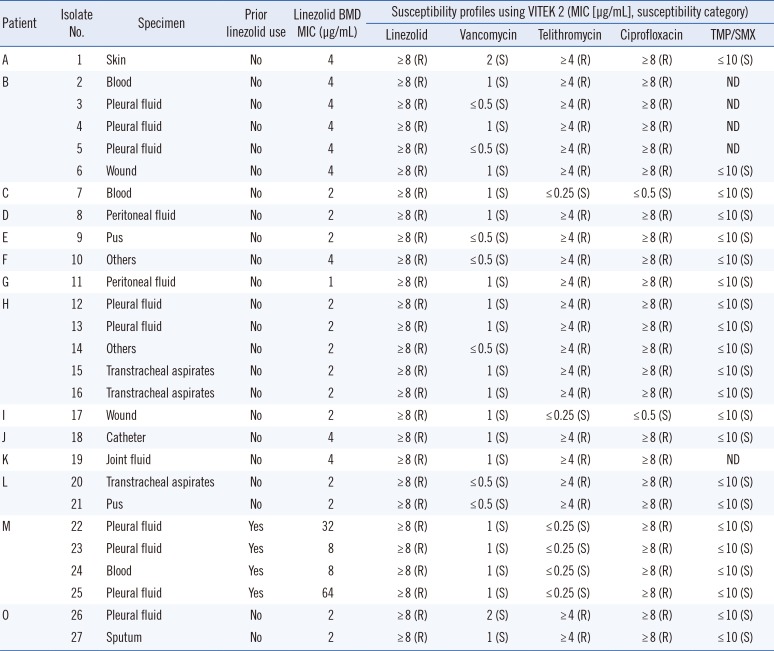
Of the 27 isolates with linezolid MIC values ≥ 8 µg/mL identified using VITEK 2, four (14.8%) isolates from one patient (Patient M) were confirmed as linezolid-resistant MRSA based on the BMD test. These four isolates had a T2500A mutation with an MIC range of 8–64 µg/mL and were resistant to ciprofloxacin but susceptible to vancomycin, telithromycin, and trimethoprim/sulfamethoxazole (
Table 2). The patient had received linezolid at the recommended dosage of 600 mg every 12 hours for 19 days. Seventeen days after the start of linezolid treatment, the first linezolid-resistant MRSA was isolated from pleural fluid (isolate 22 in
Table 3). Except for the four isolates from Patient M, the remaining 23 isolates were confirmed as linezolid-susceptible based on the BMD test, with MICs ranging from 1 to 4 µg/mL. This indicates that the linezolid-resistant results of most of the isolates (85.2%; 23/27) were major errors generated by VITEK 2. All of these isolates were susceptible to vancomycin but resistant to telithromycin and ciprofloxacin, except for two isolates that were susceptible to telithromycin and ciprofloxacin (isolates 7 and 17;
Table 2). The patients harboring these isolates had no medical history of linezolid treatment.
Table 2
Clinical characteristics and antimicrobial susceptibility profiles of 27 MRSA isolates with elevated linezolid MICs (≥8 µg/mL) using VITEK 2 (bioMérieux, Marcy-l'Étoile, France)
|
Patient |
Isolate No. |
Specimen |
Prior linezolid use |
Linezolid BMD MIC (μg/mL) |
Susceptibility profiles using VITEK 2 (MIC [μg/mL], susceptibility category) |
|
Linezolid |
Vancomycin |
Telithromycin |
Ciprofloxacin |
TMP/SMX |
|
A |
1 |
Skin |
No |
4 |
≥ 8 (R) |
2 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
B |
2 |
Blood |
No |
4 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
ND |
|
3 |
Pleural fluid |
No |
4 |
≥ 8 (R) |
≤ 0.5 (S) |
≥ 4 (R) |
≥ 8 (R) |
ND |
|
4 |
Pleural fluid |
No |
4 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
ND |
|
5 |
Pleural fluid |
No |
4 |
≥ 8 (R) |
≤ 0.5 (S) |
≥ 4 (R) |
≥ 8 (R) |
ND |
|
6 |
Wound |
No |
4 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
C |
7 |
Blood |
No |
2 |
≥ 8 (R) |
1 (S) |
≤ 0.25 (S) |
≤ 0.5 (S) |
≤ 10 (S) |
|
D |
8 |
Peritoneal fluid |
No |
2 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
E |
9 |
Pus |
No |
2 |
≥ 8 (R) |
≤ 0.5 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
F |
10 |
Others |
No |
4 |
≥ 8 (R) |
≤ 0.5 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
G |
11 |
Peritoneal fluid |
No |
1 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
H |
12 |
Pleural fluid |
No |
2 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
13 |
Pleural fluid |
No |
2 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
14 |
Others |
No |
2 |
≥ 8 (R) |
≤ 0.5 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
15 |
Transtracheal aspirates |
No |
2 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
16 |
Transtracheal aspirates |
No |
2 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
I |
17 |
Wound |
No |
2 |
≥ 8 (R) |
1 (S) |
≤ 0.25 (S) |
≤ 0.5 (S) |
≤ 10 (S) |
|
J |
18 |
Catheter |
No |
4 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
K |
19 |
Joint fluid |
No |
4 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
ND |
|
L |
20 |
Transtracheal aspirates |
No |
2 |
≥ 8 (R) |
≤ 0.5 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
21 |
Pus |
No |
2 |
≥ 8 (R) |
≤ 0.5 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
M |
22 |
Pleural fluid |
Yes |
32 |
≥ 8 (R) |
1 (S) |
≤ 0.25 (S) |
≥ 8 (R) |
≤ 10 (S) |
|
23 |
Pleural fluid |
Yes |
8 |
≥ 8 (R) |
1 (S) |
≤ 0.25 (S) |
≥ 8 (R) |
≤ 10 (S) |
|
24 |
Blood |
Yes |
8 |
≥ 8 (R) |
1 (S) |
≤ 0.25 (S) |
≥ 8 (R) |
≤ 10 (S) |
|
25 |
Pleural fluid |
Yes |
64 |
≥ 8 (R) |
1 (S) |
≤ 0.25 (S) |
≥ 8 (R) |
≤ 10 (S) |
|
O |
26 |
Pleural fluid |
No |
2 |
≥ 8 (R) |
2 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |
|
27 |
Sputum |
No |
2 |
≥ 8 (R) |
1 (S) |
≥ 4 (R) |
≥ 8 (R) |
≤ 10 (S) |

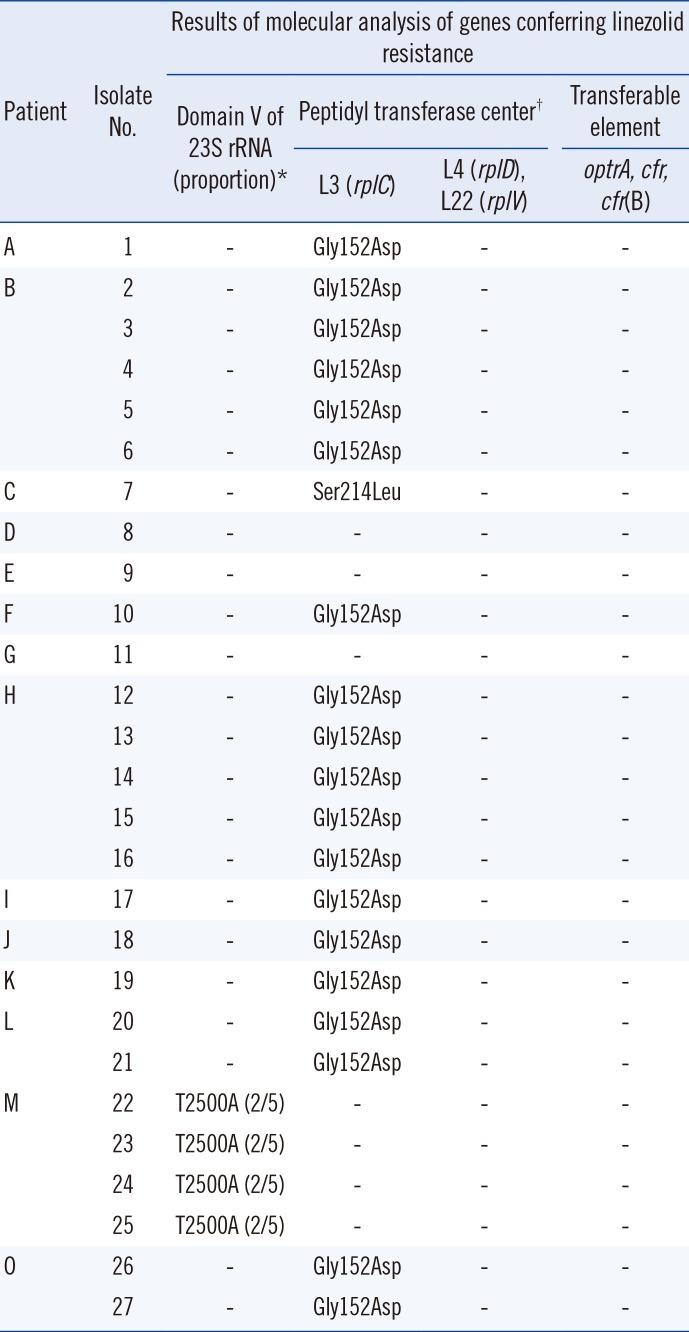
Table 3
Molecular analysis of 27 MRSA isolates with elevated linezolid MICs (≥8 µg/mL) using VITEK 2 (bioMérieux, Marcy-l'Étoile, France)
|
Patient |
Isolate No. |
Results of molecular analysis of genes conferring linezolid resistance |
|
Domain V of 23S rRNA (proportion)*
|
Peptidyl transferase center†
|
Transferable element |
|
L3 (rplC) |
L4 (rplD), L22 (rplV) |
optrA, cfr, cfr(B) |
|
A |
1 |
- |
Gly152Asp |
- |
- |
|
B |
2 |
- |
Gly152Asp |
- |
- |
|
3 |
- |
Gly152Asp |
- |
- |
|
4 |
- |
Gly152Asp |
- |
- |
|
5 |
- |
Gly152Asp |
- |
- |
|
6 |
- |
Gly152Asp |
- |
- |
|
C |
7 |
- |
Ser214Leu |
- |
- |
|
D |
8 |
- |
- |
- |
- |
|
E |
9 |
- |
- |
- |
- |
|
F |
10 |
- |
Gly152Asp |
- |
- |
|
G |
11 |
- |
- |
- |
- |
|
H |
12 |
- |
Gly152Asp |
- |
- |
|
13 |
- |
Gly152Asp |
- |
- |
|
14 |
- |
Gly152Asp |
- |
- |
|
15 |
- |
Gly152Asp |
- |
- |
|
16 |
- |
Gly152Asp |
- |
- |
|
I |
17 |
- |
Gly152Asp |
- |
- |
|
J |
18 |
- |
Gly152Asp |
- |
- |
|
K |
19 |
- |
Gly152Asp |
- |
- |
|
L |
20 |
- |
Gly152Asp |
- |
- |
|
21 |
- |
Gly152Asp |
- |
- |
|
M |
22 |
T2500A (2/5) |
- |
- |
- |
|
23 |
T2500A (2/5) |
- |
- |
- |
|
24 |
T2500A (2/5) |
- |
- |
- |
|
25 |
T2500A (2/5) |
- |
- |
- |
|
O |
26 |
- |
Gly152Asp |
- |
- |
|
27 |
- |
Gly152Asp |
- |
- |

The molecular mechanisms of linezolid resistance detected in the MRSA isolates are described in
Table 3. Analysis of domain V of the 23S rRNA gene sequences showed that the four linezolid-resistant clinical isolates from Patient M had a T to A mutation at position 2500 (T2500A;
E. coli numbering) and had two copies of the mutant 23S rRNA gene. No other mutations were detected in domain V of the 23S rRNA gene. Most (19/27; 70.4%) MRSA isolates recovered from nine patients harbored an L3 Gly152Asp mutation. However, all isolates with the Gly152Asp mutation were confirmed as linezolid-susceptible based on the BMD test. Another single isolate harbored an L3 Ser214Leu mutation. None of the isolates contained the
optrA,
cfr, or
cfr(B) genes nor any L4 or L22 protein alterations.
As clinical
S. aureus isolates exhibiting linezolid resistance are rare, few studies to date have examined acquired resistance mechanisms [
1112]. We investigated known resistance mechanisms including acquired mutations in domain V region of the 23S rRNA gene; ribosomal proteins L3, L4, and L22; and the presence of plasmid-carried genes such as
optrA,
cfr, and
cfr(B). The T2500A and G2576T mutations in the 23S rRNA gene are the most commonly identified in clinical
S. aureus [
213]. Consistent with previous studies, all linezolid-resistant MRSA isolates confirmed by the BMD test had a T2500A mutation in two copies of the gene [
2]. Linezolid resistance due to the T2500A mutation is associated with linezolid exposure rather than horizontal transmission, as a linezolid-susceptible MRSA isolate was obtained from the patient prior to linezolid exposure. In addition to mutations in linezolid binding sites caused by the ribosomal proteins, acquired resistance mechanisms due to plasmid-carried genes have been previously described in linezolid-resistant staphylococci and enterococci [
3613]. However, we did not detect any transposable elements in the studied isolates.
In addition, we detected an L3 Gly152Asp mutation in 19 MRSA isolates confirmed as linezolid-susceptible based on the BMD test. Using an
in vitro linezolid serial passage test, the MRSA-acquired L3 Gly152Asp mutation coupled with G2447T mutation in the 23S rRNA gene displayed a two- to four-fold increase in linezolid MIC compared with the G2447T mutation alone. The L3 Gly152Asp mutation probably led to a loss of oxazolidinone affinity by indirectly disrupting bases 2505 and 2506 of 23S rRNA in a manner similar to that associated with the G2576T mutation in the 23S rRNA gene [
5]. Similarly, Baos, et al. [
14] reported that the linezolid-resistant
S. epidermidis strains with a combination of the G2576T and Gly152Asp mutations showed higher linezolid MICs. However, we found only the L3 Gly152Asp mutation in clinical MRSA isolates from patients without previous exposure to linezolid, together with lower MICs (1–4 µg/mL) based on the BMD test. This mutation was not found in linezolid-susceptible MRSA isolates with lower MICs (2–4 µg/mL) using VITEK 2 (data not shown). Although the role of the L3 Gly152Asp mutation in linezolid resistance needs to be further studied, our findings demonstrated that this mutation could be acquired without linezolid exposure.
Compared with the BMD reference test, VITEK 2 determined that 23 linezolid-susceptible
S. aureus isolates were resistant, indicating that 85.2% of linezolid-resistance results using VITEK 2 were major errors. Similarly, Doern, et al. [
15] reported a poor correlation between phenotypic susceptibility testing methods, especially for detecting linezolid resistance; only 55.6% of the studied isolates generated concordant results for phenotypic methods such as VITEK 2, E-test (bioMérieux), disk diffusion, MicroScan (Dade Behring, Inc., West Sacramento, CA, USA), and the BMD test. In addition, Tenover, et al. [
16] showed poor categorical agreement between susceptibility testing methods. Our data indicate that VITEK 2 generates false resistance results; thus, specific confirmatory testing may be required.
This study had several limitations. It used relatively few isolates, which were obtained from a single institution, without performing molecular genotyping. Although a relatively small number of linezolid-resistant MRSA isolates were tested, the present data showed that a single point mutation, T2500A, in the 23S rRNA gene was mainly associated with linezolid resistance in clinical MRSA isolates from patients previously treated with linezolid. Notably, the L3 Gly152Asp mutation, most commonly detected in this study, was acquired without linezolid exposure. In addition, our results indicate that owing to the high probability of false results using VITEK 2, a confirmatory test, such as BMD test, is necessary to identify linezolid resistance in MRSA.







 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download