1. Mak LY, Cruz-Ramón V, Chinchilla-López P, Torres HA, LoConte NK, Rice JP, et al. Global epidemiology, prevention, and management of hepatocellular carcinoma. Am Soc Clin Oncol Educ Book. 2018; 38:262–279. PMID:
30231359.

2. Margini C, Dufour JF. The story of HCC in NAFLD: from epidemiology, across pathogenesis, to prevention and treatment. Liver Int. 2016; 36:317–324. PMID:
26601627.

3. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010; 7:448–458. PMID:
20628345.

4. Rastogi A. Changing role of histopathology in the diagnosis and management of hepatocellular carcinoma. World J Gastroenterol. 2018; 24:4000–4013. PMID:
30254404.

5. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018; 15:599–616. PMID:
30061739.

6. Peng JC, Shen J, Ran ZH. Transcribed ultraconserved region in human cancers. RNA Biol. 2013; 10:1771–1777. PMID:
24384562.

7. He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014; 344:20–27. PMID:
24183851.

8. Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long noncoding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014; 35:507–514. PMID:
24296588.

9. Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, et al. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014; 2014:780521. PMID:
24757675.

10. Sun J, Bie B, Zhang S, Yang J, Li Z. Long non-coding RNAs: critical players in hepatocellular carcinoma. Int J Mol Sci. 2014; 15:20434–20448. PMID:
25387074.
11. Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015; 6:7899–7917. PMID:
25760077.

12. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015; 75:3181–3191. PMID:
26069248.

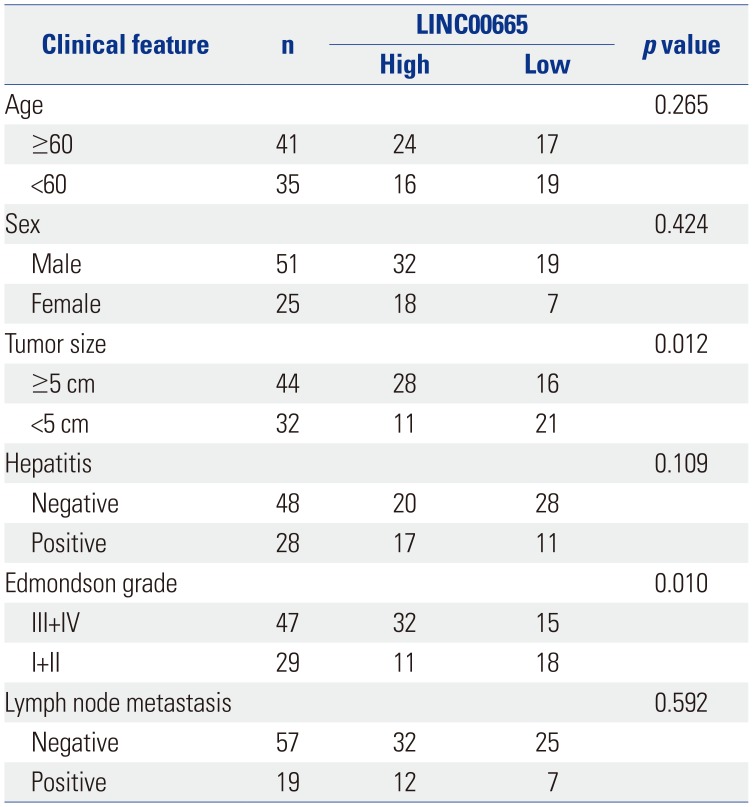
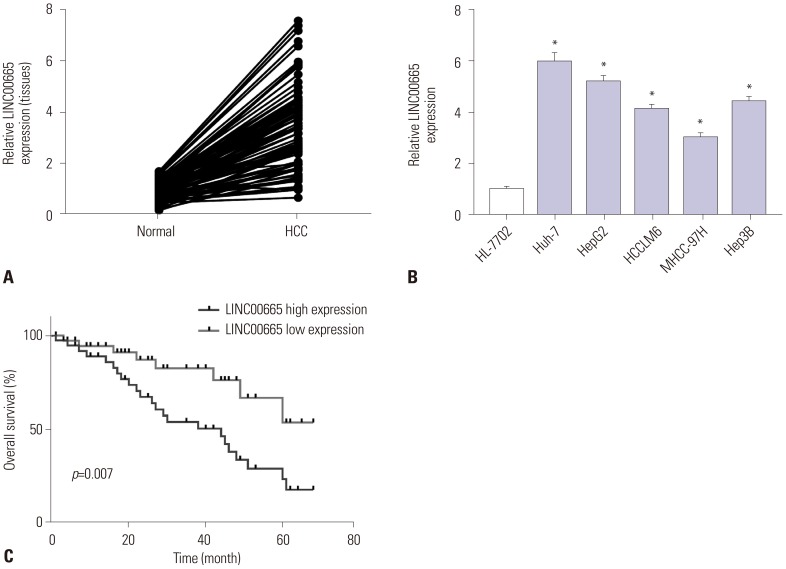
13. Wen DY, Lin P, Pang YY, Chen G, He Y, Dang YW, et al. Expression of the long intergenic non-protein coding RNA 665 (LINC00665) gene and the cell cycle in hepatocellular carcinoma using The Cancer Genome Atlas, the Gene Expression Omnibus, and Quantitative Real-Time Polymerase Chain Reaction. Med Sci Monit. 2018; 24:2786–2808. PMID:
29728556.

14. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014; 9:287–314. PMID:
24079833.

15. Song Y, Wang F, Huang Q, Cao Y, Zhao Y, Yang C. MicroRNAs contribute to hepatocellular carcinoma. Mini Rev Med Chem. 2015; 15:459–466. PMID:
25807945.

16. Guo W, Tan W, Liu S, Huang X, Lin J, Liang R, et al. MiR-570 inhibited the cell proliferation and invasion through directly targeting B7-H1 in hepatocellular carcinoma. Tumour Biol. 2015; 36:9049–9057. PMID:
26084609.

17. Oksuz Z, Serin MS, Kaplan E, Dogen A, Tezcan S, Aslan G, et al. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 2015; 42:713–720. PMID:
25391771.

18. Arbogast F, Gros F. Lymphocyte autophagy in homeostasis, activation, and inflammatory diseases. Front Immunol. 2018; 9:1801. PMID:
30127786.

19. Wang Z, Choi ME. Autophagy in kidney health and disease. Antioxid Redox Signal. 2014; 20:519–537. PMID:
23642034.

20. Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, et al. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018; 37:1142–1158. PMID:
29255248.

21. Folkerts H, Hilgendorf S, Vellenga E, Bremer E, Wiersma VR. The multifaceted role of autophagy in cancer and the microenvironment. Med Res Rev. 2019; 39:517–560. PMID:
30302772.

22. Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015; 6:8474–8490. PMID:
25893379.

23. Zhai H, Fesler A, Ju J. MicroRNA: a third dimension in autophagy. Cell Cycle. 2013; 12:246–250. PMID:
23255136.
24. Du F, Feng Y, Fang J, Yang M. MicroRNA-143 enhances chemosensitivity of Quercetin through autophagy inhibition via target GABARAPL1 in gastric cancer cells. Biomed Pharmacother. 2015; 74:169–177. PMID:
26349981.

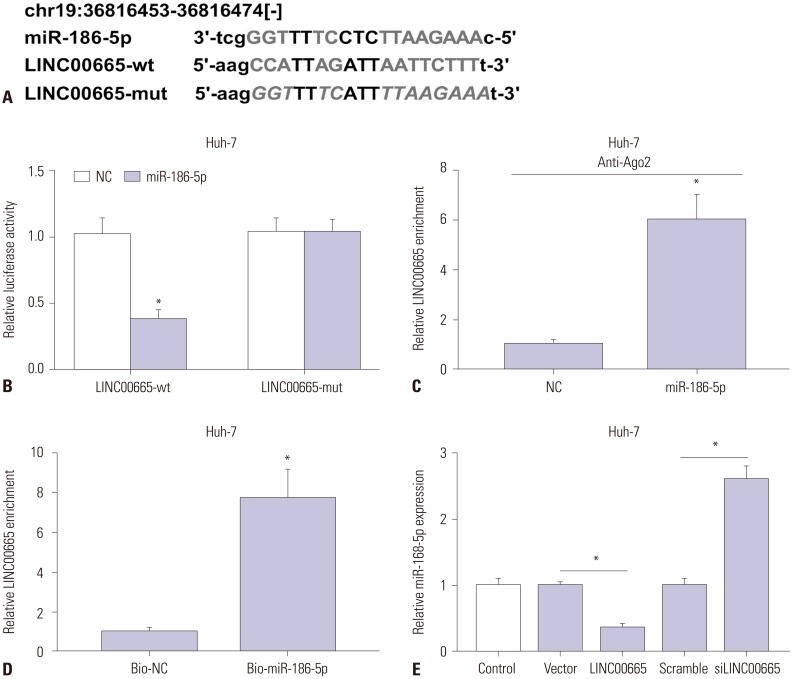
25. Lan T, Yan X, Li Z, Xu X, Mao Q, Ma W, et al. Long non-coding RNA PVT1 serves as a competing endogenous RNA for miR-186-5p to promote the tumorigenesis and metastasis of hepatocellular carcinoma. Tumour Biol. 2017; 39:1010428317705338. PMID:
28656879.

26. Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015; 36:423–434. PMID:
25968300.

27. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014; 25:666–681. PMID:
24768205.

28. Huang YH, Al-Aidaroos AQ, Yuen HF, Zhang SD, Shen HM, Rozycka E, et al. A role of autophagy in PTP4A3-driven cancer progression. Autophagy. 2014; 10:1787–1800. PMID:
25136802.

29. Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011; 18:571–580. PMID:
21311563.

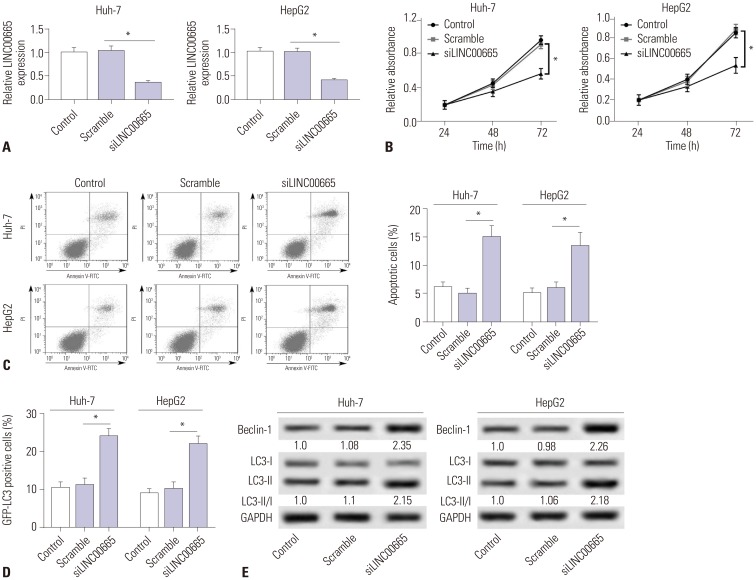
30. Qiu DM, Wang GL, Chen L, Xu YY, He S, Cao XL, et al. The expression of beclin-1, an autophagic gene, in hepatocellular carcinoma associated with clinical pathological and prognostic significance. BMC Cancer. 2014; 14:327. PMID:
24885292.

31. Hu D, Wu J, Xu L, Zhang R, Chen L. A method for the establishment of a cell line with stable expression of the GFP-LC3 reporter protein. Mol Med Rep. 2012; 6:783–786. PMID:
22825056.

32. Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H, et al. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018; 640:53–60. PMID:
29325758.

33. Islam F, Gopalan V, Vider J, Wahab R, Ebrahimi F, Lu CT, et al. MicroRNA-186-5p overexpression modulates colon cancer growth by repressing the expression of the FAM134B tumour inhibitor. Exp Cell Res. 2017; 357:260–270. PMID:
28549913.

34. Wang H, Shen Q, Zhang X, Yang C, Cui S, Sun Y, et al. The long noncoding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017; 41:2221–2229. PMID:
28448993.

35. Jones DZ, Schmidt ML, Suman S, Hobbing KR, Barve SS, Gobejishvili L, et al. Micro-RNA-186-5p inhibition attenuates proliferation, anchorage independent growth and invasion in metastatic prostate cancer cells. BMC Cancer. 2018; 18:421. PMID:
29653561.

36. Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011; 75:50–83. PMID:
21372320.

37. Hsu C, Lee EX, Gordon KL, Paz EA, Shen WC, Ohnishi K, et al. MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat Commun. 2018; 9:942. PMID:
29507340.

38. Liu L, Lu L, Zheng A, Xie J, Xue Q, Wang F, et al. MiR-199a-5p and let-7c cooperatively inhibit migration and invasion by targeting MAP4K3 in hepatocellular carcinoma. Oncotarget. 2017; 8:13666–13677. PMID:
28099144.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download