Abstract
Although sciences and technology have progressed rapidly, de novo drug development has been a costly and time-consuming process over the past decades. In view of these circumstances, ‘drug repurposing’ (or ‘drug repositioning’) has appeared as an alternative tool to accelerate drug development process by seeking new indications for already approved drugs rather than discovering de novo drug compounds, nowadays accounting for 30% of newly marked drugs in the U.S. In the meantime, the explosive and large-scale growth of molecular, genomic and phenotypic data of pharmacological compounds is enabling the development of new area of drug repurposing called computational drug repurposing. This review provides an overview of recent progress in the area of computational drug repurposing. First, it summarizes available repositioning strategies, followed by computational methods commonly used. Then, it describes validation techniques for repurposing studies. Finally, it concludes by discussing the remaining challenges in computational repurposing.
De novo drug development is an expensive and time-consuming process. It is known that the total average development cost for a new drug ranges from $2 to $3 billion and the total development time takes at least 13–15 years.[1] Further, it suffers from a high attrition rate. Of the drugs entering phase 1 clinical trials, only 10% are approved, the rest failing due to high toxicity or inefficacy.[2] These attritions are mainly due to inaccurate identification of the drug target or response. Despite rapid advancements in technologies and geometric increases in R&D spending, the number of drugs newly approved remains the same.[3] Moreover, in the oncology area only 5% of drug compounds entering Phase I clinical trials are approved,[4] and in the area of orphan drugs, more than 8000 related diseases exist, making the de novo drug development impossible for this huge number of diseases with the current R&D costs.[5]
Within this context, finding new indications and targets for already marketed drugs, an approach called ‘drug repurposing’ (or ‘drug repositioning’), which was first discussed by Ashburn and Thor in 2004,[2] has begun to fill the gap for the lack of efficiency of the traditional drug development.[6] The major advantage of drug-repurposing approaches is that, for an existing drug, not only preclinical information but also clinical profiles (pharmacokinetic, pharmacodynamic and toxicity) are already available, thereby reducing the development risk. Accordingly, the drug compound can rapidly enter late-stage clinical trials, reducing development cost and time.[7] Therefore, it is not surprising that nowadays about 30% of newly approved drugs are repositioned drugs in the U.S.[2] Table 1 lists some of the repurposed drugs developed so far, and Figure 1 conceptually displays de novo drug development and drug repurposing processes.
Drug repurposing is performed either experimentally or computationally. The latter approach is also called ‘in silico drug repurposing’,[8] which belongs to the area of computational pharmacology. In silico drug repurposing is classified into discovering new indications for an existing drug (drug-centric) and identifying effective drugs for a disease (disease-centric) and has the common strategy of similarity assessment between drugs and/or diseases.[9] Various computational repurposing approaches were reviewed in Jin and Wong.[10]
The development of in silico drug repurposing and its wide use today have been made possible by the following two technological trends.[8] The first trend is that high-throughput data from various sources, including genomics, proteomics, chemo-proteomics, and phenomics, have been generated and accumulated. As a result, not only data characterizing disease phenotypes and drug profiles, but entire pathway maps have become available. The second is that, due to the advances in computational and data sciences, it has been possible for repurposing algorithms to develop, along with retrospective analysis and database maintenance for experimental data.[11]
In this repurposing strategy, utilizing the drug-related information, including drug targets, chemical structures, pathways, adverse effects, etc., models are built to predict unknown targets, bio-markers or mechanisms for diseases.[12] This strategy includes target-based, pathway-based, and target mechanismbased drug-repurposing.
Given proteins or biomarkers of interest, target-based drug-repurposing comprises high-throughput and/or high-content screening (HTS/HCS) of drug compounds,[13] followed by in silico screening of drug compounds from drug libraries, such as ligand-based screening or docking.[14] Compared with blinded search or screening which does not use biological or pharmacological information when screening, target-based repurposing directly links targets with disease mechanisms and therefore the likelihood of drug discovery significantly improves. The advantage of the target-based approach lies in its ability to screen nearly all drug compounds with known chemical structure. However, target-based methods cannot identify unknown mechanisms beyond the targets already known.
Pathway-based drug-repurposing utilizes metabolic pathways, signaling pathways, and protein-interaction networks information to predict the similarity or connection between disease and drug. For example, using omics data processed from human patients or animals, disease-specific pathways are reconstructed to serve as new targets for repositioned drugs.[15]
Target mechanism-based drug-repurposing integrates signaling pathway information, treatment omics data, and protein interaction networks to discover new mechanisms of action for drugs.[16] The necessity of precision medicine, which has been increasingly important, motivates such drug-repurposing approaches. The advantage of these repurposing approaches is that they aim to discover the mechanisms related not only to diseases or drugs but also to drug treatments to specific diseases.
In signature-based repurposing, gene signatures information obtained from disease omics data[17] is used to discover new off-targets or mechanisms of disease. This approach searches inverse drug–disease relationships by comparing gene expression profiles between drug and disease. In the work by Dudley et al.,[20] potential drug–disease pairs were investigated for inflammatory bowel disease (IBD), where gene expression profiles obtained from the gene expression omnibus database[18] were compared with gene expression profiles comprising 164 drug compounds obtained from the connectivity map.[19] As a result, unknown drug–disease pairs were discovered, with one pair validated in preclinical models.
The advantage of these approaches is that they identify new mechanisms of action for drugs. Also, unlike knowledge-based methods, more molecular- and/or genetic-level mechanisms are involved in these methods.
The phenotypic information has become available as a new source of drug repositioning. In recent years, this type of information has been increasingly used by systems approaches to detect genetic traits associated with human diseases.[21] Natural language processing skills applied to electronic health records (EHRs) can reveal additional adverse drug events which were not observed during drug development.[22] For example, mining EHRs helped in identifying that metformin can be repurposed for cancer treatment.[23]
Machine learning (ML) techniques that have been applied for drug repositioning include logistic regression, support vector machine (SVM), random forest, neural network (NN), and deep learning (DL).
For logistic regression, PREDICT, a similarity-based ML framework, has been reported where drug-drug similarity was integrated with disease-disease similarity and integrated similarity values were used as features in predicting similar drugs for similar diseases using logistic regression.[24] SPACE, another similarity-based ML method, also used logistic regression to predict the therapeutic chemical class of a drug by integrating multiple sources of data.[25]
For SVM, Napolitano et al.[26] predicted drug therapeutic class by a SVM approach based on molecular target, drug chemical structure, and gene expression similarity. In their work, these features were merged into a single drug similarity matrix to be used as a kernel for SVM classification. Similarly, Wang et al.[27] proposed a SVM model incorporating molecular activity, drug chemical structure, and side effect. Three types of data were then integrated to construct a kernel function of SVM classifier, and their method showed higher efficiency than other methods.
For NN, Menden et al. [28] developed a NN-based prediction model for cancer cell line response to drug treatment, parameterized by IC50. In their model, genomic (e.g., microsatellite status and mutation status of 77 oncogenes) and chemical features (e.g., structural fingerprint) of cancer cell lines were analyzed to build a perceptron NN and random forest regression.
DL, when compared to shallow learning, is capable of discovering latent and complex structure in large datasets and, by using backpropagation algorithms, allows adjusting connecting weights as well, enabling to compute the representation of each layer based on that of the previous layer.[29] Aliper and Plis[30] analyzed gene expression profile data using a DL approach to predict therapeutic categories of drugs and found deep neural networks (DNN) surpassed SVM, suggesting evidence for applying DL to drug development as a useful tool. Additional reports suggested that by multi-task learning DL-based approaches outperform traditional ML algorithms in predicting the toxicity.[31]
In these models, network nodes represent drug, disease, or gene products while edges represent interactions or relationships between nodes. Networks are knowledge-based or computationally inferred from multiple data sources and represent various interactions, including drug-drug, drug-target, drugdisease, disease-disease, disease-gene, and protein-protein interactions, and transcriptional and signaling networks.[32] Based on the ‘guilt-by-association’ principle, by integrating heterogeneous data, the method can discover unknown or hidden drug-disease relationships. According to this principle, drugs provoking similar transcriptional responses could have a similar mode of action.[33]
Using a bi-partite network, Cheng and Liu[34] compared similarities between drug-based, target-based, and network-based interactions and used them in predicting drug-target interaction, finding that network-based inference showed the best performance as compared by the area under receiver operating curve (AUROC). Also, using a drug–disease heterogeneous network model, Wu et al.[35] identified closely connected modules of drugs and diseases, so as to extract the information on potential drug–disease pair candidates for drug repositioning. Jin et al.[17] proposed a repurposing method for cancer drugs by leveraging potential off-target effects on cancer cell signaling pathways.
The biomedical and pharmaceutical literature contains a huge amount of information available for drugs and diseases, from which potential indications for existing drugs can be detected through text mining schemes.[36] One basis for this scheme is biological ontology, which enables to compare and analyze biological information from various sources. Several text mining approaches for drug repurposing are summarized in a recent study[37]; for example, if nutrition B deficiency was found to cause disease A in one study while another study found drug C to be an activator of nutrition B in another disease, then text mining would recommend repurposing drug C for disease A.
Semantic inference involves technologies such as topic modeling that facilitate the discovery of drug indications by integrating various data sources. For instance, Bisgin et al.,[38] proposed Latent Dirichlet Allocation based drug repositioning incorporating a topic model to process the phenome information for drug side effects. Also, by modeling relations between breast cancer drugs approved by FDA and associated genes, pathways, SNPs and diseases, Zhu et al.,[39] developed an ontology-based knowledge tool to predict potential disease-drug pairs in breast cancer. Chen et al.,[40] proposed a semantic linked network-based approach to assess drug–target associations, which comprised drugs, protein targets, chemical compounds, diseases, pathways, side effects, and their relations. In their model, the topology and semantics were represented by the subgraph between a drug and a target, where drug–target pairs located in different disease areas in the model but showing the similarity were associated with a potential repositioning opportunity.
Validation of repurposing results can be done computationally and experimentally. For computational validation, a straightforward way is to assess AUROC values as well as specificity, sensitivity and positive predictive value (PPV).[41] In addition, recall and precision can be computed to obtain the precision-recall curve (PRC),[42] followed by the area under this curve (AUPRC).[43] In addition, the validity can be assessed by comparing predicted targets in PubMed, ClinicalTrials or EHRs. For example, EHRs were used to validate the metformin effects associated with cancer mortality.[24]
For experimental validation, cell-based targeted assays (in vitro and in vivo) and animal experiments need to be performed. For example, atorvastatin's beneficial effect on graft survival was observed in a single-center cohort of 2,515 patients receiving renal transplantation by retrospective analysis of EHRs followed for up to 22 years, which was validated in a meta-analysis using public microarray datasets where atorvastatin was also found to be beneficial for organ transplantation.[44]
Computational drug repurposing greatly reduces drug development costs and time by discovering new indications for existing drugs. This method enables the joint analysis of different sources of data, including genomic, biomedical and pharmacological data, which improves drug repositioning efficiency.
In this review, available repositioning methods were described according to the source of data and information used. With the increased importance of precision medicine and personalized drug, mechanism-based repurposing approaches are expected to be extended to finding new indications for individual patients as these repurposing approaches can take into account patients heterogeneity and complexity, reducing the risk of drug toxicity or inefficacy caused by inter-patient variability.[45]
A few issues to consider in computational repurposing are as follows. First, repurposing results are sensitive to datasets used, more reliable results obtained with more sources of data. Second, although more studies are needed, previous works show DNN-based repurposing methods outperform other ML-based methods such as SVM or random forest. Given that DL methods have a good performance in pattern recognition such as image and configuration, it may be possible to use recursive or convolutional neural networks to assess the potential toxicity of drug compounds based on raw structures.[46] DL-based methods have been reported to be effective for assessing drug toxicity.[32]
Figures and Tables
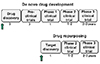 | Figure 1Conceptual diagram of de novo drug development and drug repurposing; the arrow denotes the initiation time in each scenario and the number denotes a period of time in years. In this figure, the objective of drug repurposing was assumed to be to identify or discover new targets for a drug marketed. Note that drug repurposing begins with target discovery for an existing drug, directly followed by phase 2 and 3 clinical trials, while animal and phase 1 clinical studies were not conducted as results for these studies are already available for an existing drug. |
Table 1
Examples of repositioned drugs (selected)

References
1. Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012; 11:191–200. DOI: 10.1038/nrd3681.

2. Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013; 12:581–594. DOI: 10.1038/nrd4051.

3. Booth B, Zemmel R. Opinion/Outlook: Prospects for productivity. Nat Rev Drug Discov. 2004; 3:451–456.
4. Kato S, Moulder SL, Ueno NT, Wheler JJ, Meric-Bernstam F, Kurzrock R, et al. Challenges and perspective of drug repurposing strategies in early phase clinical trials. Oncoscience. 2015; 2:576–580.

5. Sardana D, Zhu C, Zhang M, Gudivada RC, Yang L, Jegga AG. Drug repositioning for orphan diseases. Brief Bioinform. 2011; 12:346–356. DOI: 10.1093/bib/bbr021.

6. Munos BH, Chin WW. How to revive breakthrough innovation in the pharmaceutical industry. Sci Transl Med. 2011; 3:89cm16. DOI: 10.1126/scitranslmed.3002273.

7. Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013; 34:267–272. DOI: 10.1016/j.tips.2013.03.004.

8. Shim JS, Liu JO. Recent advances in drug repositioning for the discovery of new anticancer drugs. Int J Biol Sci. 2014; 10:654–663. DOI: 10.7150/ijbs.9224.

9. Liu Z, Fang H, Reagan K, Xu X, Mendrick DL, Slikker W Jr, et al. In silico drug repositioning: What we need to know. Drug Discov Today. 2013; 18:110–115. DOI: 10.1016/j.drudis.2012.08.005.
10. Jin G, Wong ST. Toward better drug repositioning: Prioritizing and integrating existing methods into efficient pipelines. Drug Discov Today. 2014; 19:637–644. DOI: 10.1016/j.drudis.2013.11.005.

11. Hodos RA, Kidd BA, Shameer K, Readhead BP, Dudley JT. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip Rev Syst Biol Med. 2016; 8:186–210. DOI: 10.1002/wsbm.1337.
12. Emig D, Ivliev A, Pustovalova O, Lancashire L, Bureeva S, Nikolsky Y, et al. Drug Target Prediction and Repositioning Using an Integrated Network-Based Approach. PLoS ONE. 2013; 8:e60618. DOI: 10.1371/journal.pone.0060618.

13. Swamidass SJ. Mining small-molecule screens to repurpose drugs. Brief Bioinform. 2011; 12:327–335. DOI: 10.1093/bib/bbr028.

14. Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, et al. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem. 2002; 45:2213–2221.

15. Jadamba E, Shin M. A Systematic Framework for Drug Repositioning from Integrated Omics and Drug Phenotype Profiles Using Pathway-Drug Network. BioMed Res Int. 2016; 2016:7147039. DOI: 10.1155/2016/7147039.

16. Jin G, Fu C, Zhao H, Cui K, Chang J, Wong ST. A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res. 2012; 72:33–44. DOI: 10.1158/0008-5472.CAN-11-2333.

17. Haeberle H, Dudley JT, Liu JT, Butte AJ, Contag CH. Identification of cell surface targets through meta-analysis of microarray data. Neoplasia. 2012; 14:666–669.

18. Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, et al. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005; 33:D562–D566.

19. Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006; 313:1929–1935.

20. Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011; 3:96ra76. DOI: 10.1126/scitranslmed.3002648.

21. Hebbring SJ. The challenges, advantages and future of phenome-wide association studies. Immunology. 2014; 141:157–165. DOI: 10.1111/imm.12195.

22. Luo Y, Thompson WK, Herr TM, Zeng Z, Berendsen MA, Jonnalagadda SR, et al. Natural Language Processing for EHR-Based Pharmacovigilance: A Structured Review. Drug Saf. 2017; 40:1075–1089. DOI: 10.1007/s40264-017-0558-6.

23. Xu H, Aldrich MC, Chen Q, Liu H, Peterson NB, Dai Q, et al. Validating drug repurposing signals using electronic health records: A case study of metformin associated with reduced cancer mortality. J Am Med Inform Assoc. 2015; 22:179–191. DOI: 10.1136/amiajnl-2014-002649.

24. Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: A method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011; 7:496. DOI: 10.1038/msb.2011.26.

25. Liu Z, Guo F, Gu J, Wang Y, Li Y, Wang D, et al. Similarity-based prediction for Anatomical Therapeutic Chemical classification of drugs by integrating multiple data sources. Bioinformatics. 2015; 31:1788–1795. DOI: 10.1093/bioinformatics/btv055.

26. Napolitano F, Zhao Y, Moreira VM, Tagliaferri R, Kere J, D'Amato M, et al. Drug repositioning: a machine-learning approach through data integration. J Cheminform. 2013; 5:30. DOI: 10.1186/1758-2946-5-30.

27. Wang Y, Chen S, Deng N, Wang Y. Drug repositioning by kernel based integration of molecular structure, molecular activity, and phenotype data. PLoS One. 2013; 8:e78518. DOI: 10.1371/journal.pone.0078518.

28. Menden MP, Iorio F, Garnett M, McDermott U, Benes CH, Ballester PJ, et al. Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLoS One. 2013; 8:e61318. DOI: 10.1371/journal.pone.0061318.

30. Aliper A, Plis S, Artemov A, Ulloa A, Mamoshina P, Zhavoronkov A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol Pharm. 2016; 13:2524–2530. DOI: 10.1021/acs.molpharmaceut.6b00248.

31. Unterthiner T, Mayr A, Klambauer G, Hochreiter S. Toxicity Prediction using Deep Learning. Front Environ Sci. 2015; 3:10.
32. Azuaje F. Drug interaction networks: An introduction to translational and clinical applications. Cardiovasc Res. 2013; 97:631–641. DOI: 10.1093/cvr/cvs289.

33. Iorio F, Rittman T, Ge H, Menden M, Saez-Rodriguez J. Transcriptional data: a new gateway to drug repositioning? Drug Discov Today. 2013; 18:350–357. DOI: 10.1016/j.drudis.2012.07.014.

34. Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G, et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol. 2012; 8:e1002503. DOI: 10.1371/journal.pcbi.1002503.

35. Wu C, Gudivada RC, Aronow BJ, Jegga AG. Computational drug repositioning through heterogeneous network clustering. BMC Syst Biol. 2013; 7 Suppl 5:S6. DOI: 10.1186/1752-0509-7-S5-S6.

36. Tari LB, Patel JH. Systematic drug repurposing through text mining. Methods Mol Biol. 2014; 1159:253–267. DOI: 10.1007/978-1-4939-0709-0_14.

37. Andronis C, Sharma A, Virvilis V, Deftereos S, Persidis A. Literature mining, ontologies and information visualization for drug repurposing. Brief Bioinform. 2011; 12:357–368. DOI: 10.1093/bib/bbr005.

38. Bisgin H, Liu Z, Fang H, Kelly R, Xu X, Tong W. A phenome-guided drug repositioning through a latent variable model. BMC Bioinformatics. 2014; 15:267.

39. Zhu Q, Tao C, Shen F, Chute CG. Exploring the pharmacogenomics knowledge base (PharmGKB) for repositioning breast cancer drugs by leveraging Web ontology language (OWL) and cheminformatics approaches. Pac Symp Biocomput. 2014; 172–182.

40. Chen B, Ding Y, Wild DJ. Assessing drug target association using semantic linked data. PLoS Comput Biol. 2012; 8:e1002574. DOI: 10.1371/journal.pcbi.1002574.

41. Guney E, Menche J, Vidal M, Barábasi AL. Network-based in silico drug efficacy screening. Nat Commun. 2016; 7:10331. DOI: 10.1038/ncomms10331.

42. Alaimo S, Giugno R, Pulvirenti A. Recommendation techniques for drug–target interaction prediction and drug repositioning. Methods Mol Biol. 2016; 1415:441–462. DOI: 10.1007/978-1-4939-3572-7_23.

43. Mei JP, Kwoh CK, Yang P, Li XL, Zheng J. Drug-target interaction prediction by learning from local information and neighbors. Bioinformatics. 2013; 29:238–245. DOI: 10.1093/bioinformatics/bts670.

44. Khatri P, Roedder S, Kimura N, De Vusser K, Morgan AA, Gong Y, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013; 210:2205–2221. DOI: 10.1084/jem.20122709.

45. Li YY, Jones SJ. Drug repositioning for personalized medicine. Genome Med. 2012; 4:27. DOI: 10.1186/gm326.

46. Xu Y, Dai Z, Chen F, Gao S, Pei J, Lai L. Deep learning for drug-induced liver injury. J Chem Inf Model. 2015; 55:2085–2093. DOI: 10.1021/acs.jcim.5b00238.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download