Abstract
Purpose
The purpose of this study was to explore the potential early diagnostic value of serum microRNA-381(miRNA-381) in patients with gastric cancer (GC).
Materials and Methods
Patients with advanced gastric cancer (AGC) and early gastric cancer (EGC), as well as healthy individuals, were enrolled in this study. Expression of miRNA-381 in serum was detected using real-time quantitative PCR. Electrochemiluminescence analysis was used to investigate the expression of classic tumor markers, including carbohydrate antigen (CA) 199, CA724, and carcinoembryonic antigen. Finally, receiver operating characteristic curve and Kaplan-Meier analysis were used to determine the value of miRNA-381 in clinical diagnosis of GC.
Results
miRNA-381 was differentially expressed among the study groups. AUC analysis showed that the sensitivity and specificity of serum miRNA-381 in the diagnosis of GC were superior to those of other tumor markers. Furthermore, low levels of miRNA-381 expression were positively correlated with lymph node metastasis and AGC. Finally, Kaplan-Meier survival analysis showed that down-regulation of miRNA-381 was associated with lymph node metastasis and the development of GC.
Gastric cancer (GC) develops from the lining of the stomach and ranks fourth in incidence and second in mortality among all cancers worldwide.12 Since most cases are diagnosed at advanced stages with poor prognosis and limited treatment options, GC remains a major clinical challenge.34 Although considerable effort has been directed toward the development of surgical and chemotherapeutic interventions, as well as increased GC screening rates, the prognosis of GC remains poor, and the molecular mechanisms of GC progression are not completely understood.567
The development of GC is a complex and multifactorial process involving a number of genetic alterations.8 During this process, the differential expression of particular genes and microRNA (miRNA) plays a very important role.9 Accumulating evidence suggests that dysregulation of miRNAs is intimately involved in the carcinogenesis, progression, and metastasis of many cancers, including GC and that the alteration of certain miRNAs may be biomarkers of use in detecting early GC.10 As a miRNA, miRNA-381 has been reported to play suppressive or promoting roles in the progression of cancer.11 Liang, et al.12 indicated that the down-regulation of miRNA-381 promotes cell proliferation and invasion in colon cancer. A previous study showed that miRNA-381 inhibits the metastasis of GC by targeting TMEM16A expression.13 Zhang, et al.14 showed that miRNA-381 inhibited migration and invasion in human GC through the down-regulation of SRY-Box 4. Although differential expression of miRNA-381 in GC has been described in previous study, the detail diagnostic value of miRNA-381 in serum in GC has not been fully investigated yet.
In the present study, we aimed to investigate the role of serum miRNA-381 in the early diagnosis of GC. We found that the levels of miRNA-381 in advanced gastric cancer (AGC) were significantly lower than those in early gastric cancer (EGC) and that reduced expression of miR-381 was positively correlated with lymph node metastasis and GC progression. We sought to explore the early diagnostic value of serum miRNA-381 in GC and to provide a novel strategy for GC therapy.
Patients with GC confirmed by gastroscopy or surgical pathology were selected from department of gastroenterology of The 5th People's Hospital of Ji'nan for May 2016 to May 2018. All GC patients were diagnosed by pathology without any antineoplastic treatment. Among them, a total of 80 serum samples were collected from AGC patients (51 males and 29 females; average age of 61.73±10.63 years). Meanwhile, 40 serum samples from EGC patients (26 males and 14 females; average age of 58.25±9.67 years) and 40 serum samples from healthy controls (22 males and 18 females; average age of 60.20±10.70 years) were collected among outpatient health examinees of People's Hospital Affiliated to Inner Mongolia Medical University. The detail baseline information of the GC patients is showed in Table 1. The experimental scheme of this study was approved by the Ethics Committee of The 5th People's Hospital of Ji'nan (IRB No. 2018066). Patient participation was voluntary, and all patients provided written informed consent.
Total RNA was extracted from early morning fasting venous blood using miRcute Serum/Plasma miRNA Isolation Kits (Beijing Tiangen Biochemical Technology Co., Ltd., Beijing, China). Reverse transcription was performed using miRNA RT Reaction Buffer and miRNA RT Enzyme Mix (Beijing Tiangen Biochemical Technology Co., Ltd.). Real-time quantitative PCR (RT-qPCR) was performed on ABI7500 (Thermo Fisher Scientific, Waltham, MA, USA) using special primers (miRNA-381, forward: 5′-UAUACAAGGGCAAGCUCUCUGU-3′; reverse: 5′-AGAGAGCUUGCCCUUGUAUAUU-3′). U6 was used as an internal control (U6, forward: 5′-TCGCCCTTGGCA CAGCA-3′; reverse: 5′-CGAACCATTCAAGTGTTGCT-3′). The PCR program included 95℃ for 10 min, 40 cycles of 95℃ for 10 s, 60℃ for 20 s, and 72℃ for 34 s. The relative expression of miRNA-381 was calculated using the 2−ΔΔCt method.15 The RT-qPCR was conducted according to the instructions of miRcute Plus miRNA qPCR Detection Kits (Thermo Fisher Scientific) and repeated three times for each experiment.
Electrochemiluminescence is chemiluminescence triggered by electrochemical techniques.16 Serum carbohydrate antigen (CA) 724, CA199, and carcinoembryonic antigen (CEA) were measured using the electrochemiluminescence immuno-assay (Roche cobas e601, Roche Diagnostics GmbH, Mannheim, Germany). The detection thresholds of CA724, CEA, and CA199 were 6.9 U/mL, 6.5 ng/mL, and 27 U/mL, respectively.
To reveal the prognostic value of factors of interest in patients with GC, survival analysis was performed. Patients with GC were grouped according to clinical information and pathological parameters, including age, sex, stage of GC, expression levels of miRNA-381, lymph node metastasis, and differentiation of tumor tissue. Survival rate estimation was performed using the Kaplan-Meier method17 and Cox proportional hazards models.1819
Statistical analysis was performed using SPSS, version 17.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad Softward, San Diego, CA, USA). The chi-square test was used to compare the counting data between groups. The measurement data are expressed as a mean±SD. The means were compared using Student's t-test between groups. One-way ANOVA followed by Tukey's multiple comparisons test was used for comparison among groups. The area under receiver operating characteristic curves (AUC) was used to analyze the clinical diagnostic value of the detection of miRNA-381. The survival time of GC patients was tested by the Kaplan-Meier method. Prognosis was analyzed by Cox regression. p<0.05 was regarded as statistically significant.
Normality test results showed that the data of each group conformed to normal distribution; therefore, the data in each group are expressed as means±SDs. The serum levels of miRNA-381 in the EGC group were significantly lower than those in the healthy group, while the levels of miRNA-381 in the AGC group were significantly lower than those in the EGC group (all p<0.05) (Fig. 1A). Moreover, the serum levels of CA199 in the EGC group were significantly higher than those in the healthy group, while the serum levels of CA199 in the AGC group were significantly higher than those in the EGC group (all p<0.05) (Fig. 1B). Furthermore, the levels of CA724 and CEA in the AGC group were significantly higher than those in the EGC group and healthy group (all p<0.05) (Fig. 1C and D).
To evaluate the value of miRNA-381 in the diagnosis of GC, receiver operating characteristic curves of serum miRNA-381, CA199, CA724, and CEA were investigated among the study groups. We found that AUC value of miRNA-381 was larger than AUC values for other tumor markers comparing the EGC group versus the healthy group (AUC of miRNA-381: 0.931) (Fig. 2A) and the EGC versus the AGC group (AUC of miRNA-381: 0.922) (Fig. 2B). The sensitivity and specificity of serum miRNA-381 in the diagnosis of GC were better than those for other tumor markers (Table 2).
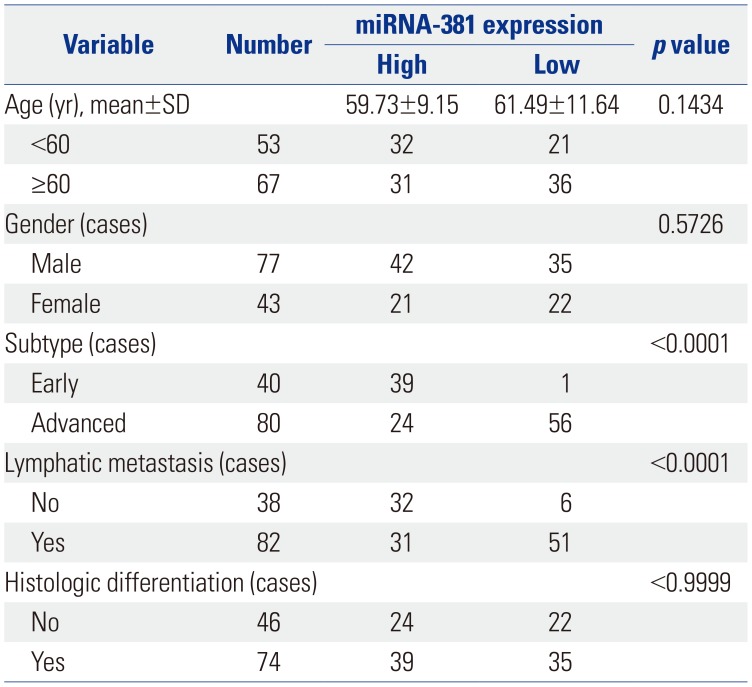
To further reveal the relationship between the expression of miRNA-381 and clinicopathological parameters of GC patients, the mean expression level of miRNA-381 (0.6173) was measured as a node. Meanwhile, a total of 120 GC patients was divided into two groups: high miRNA-381 expression group (n=63) and low miRNA-381 expression group (n=57). In doing so, we found that low expression of miRNA-381 was positively correlated with lymph node metastasis and AGC (p<0.01). Detailed information of miRNA-381 expression and associated clinical characteristics is showed in Table 3.
Kaplan-Meier survival analysis showed that the overall survival rate of GC patients with low expression of miRNA-381 was significantly shorter than that of GC patients with high expression of miRNA-381 (Fig. 3). Cox regression analysis showed that the expression of miRNA-381 [95% confidence interval (CI): 1.117–3.217; p=0.009], lymph node metastasis (95% CI: 0.295–0.828; p=0.007), and histologic differentiation (95% CI: 1.416–4.085; p=0.001) were correlated with the prognosis of GC (data not shown).
GC is one of the leading causes of cancer-related deaths worldwide with poor diagnosis and few treatment strategies.20 The current study explored the early diagnostic value of serum miRNA-381 in patients with GC. Our results showed that miRNA-381 was differentially expressed in not only EGC versus AGC but also EGC versus healthy individuals. Moreover, AUC analysis showed that the sensitivity and specificity of serum miRNA-381 in the diagnosis of GC were superior to other tumor markers. Furthermore, low levels of miRNA-381 expression were positively correlated with lymph node metastasis and AGC. Finally, Kaplan-Meier survival analysis showed that the deregulation of miRNA-381 was associated with the development of GC.
The down-regulation of miRNA-381 is closed related with the progression of cancer.21 A previous study showed that miRNA-381 is significantly down-regulated in metastatic cells, compared to normal prostatic epithelial cells.22 Liang, et al.12 indicated that the down-regulation of miRNA-381 promotes cell proliferation and invasion in colon cancer. Rothschild, et al.23 showed that miR-381 was significantly down-regulated in human lung adenocarcinomas and that low miR-381 expression levels are correlated with poor prognosis. A clinical investigation on osteosarcoma indicated that osteosarcoma patients with low expression of miRNA-381 experienced a longer survival time after surgical intervention and that miRNA-381 expression promotes cell proliferation and cell invasion ability.24 A previous study showed that the down-regulation of miRNA-148a is associated with lymph node metastasis and poor clinical outcomes and functions as a suppressor of tumor metastasis in non-small cell lung cancer.25 Fujino, et al.26 indicated that the down-regulation of miRNA-100 is associated with lymph node metastasis in early colorectal cancer with submucosal invasion. In the current study, the analysis of serum miRNA-381expression in GC patients showed that, compared with healthy controls, miRNA-381 was down-regulated in both AGC patients and EGC patients. Meanwhile, investigation of the relationship between miRNA-381 expression and clinicopathological features revealed that low expression of miRNA-381 was positive correlated with lymph node metastasis and AGC.
CEA, CA199, and CA724 are three classic biomarkers of the progression of GC.27 Ucar, et al.28 proved the prognostic values of preoperative CEA, CA199, and CA724 in GC clinical treatment. A previous study indicated that CEA and CA199 can be used for postoperative monitoring of recurrence in patients with AGC.29 Meanwhile, another study reported that CA724 (47.7%) showed a higher positivity rate for GC than CEA (25%) and CA 199 (25%).30 Compared with cystic fluid CEA determination, miRNA detection holds great promise as molecular diagnostic tools for assessing cancer risk,31 as miRNAs act as oncogenes or tumor suppressors in a wide variety of human cancers.32 A previous study suggested that the down-regulation of serum miRNA-195 is an optimal biomarker in the clinical diagnosis of breast cancer, compared to CEA and CA153.33 Zhang, et al.34 indicated that miRNA-181a functions as an biomarker in GC by targeting certain tumor suppressor genes. Moreover, genome-wide miRNA profiles have suggested miR-378 as a serum biomarker for early detection of GC.35 In the current study, AUC analysis based on potential tumor markers, including miRNA-381, CA199, CA724, and CEA showed that the sensitivity and specificity of serum miRNA-381 in the diagnosis of GC were superior to the other tumor markers.
In conclusion, we suggest that miRNA-381, which is down-regulated in GC, might be a novel early diagnosis marker for patients with GC. Furthermore, we discovered that the down-regulation of miRNA-381 is positively correlated with lymph node metastasis and advanced stage disease in GC.
Notes
AUTHOR CONTRIBUTIONS:
Conceptualization: Ye Li.
Data curation: Ye Li.
Formal analysis: Ye Li.
Investigation: Huihui Sun.
Methodology: Jie Guan.
Project administration: Jie Guan.
Resources: Tingting Ji.
Software: Tingting Ji.
Supervision: Ye Li, Huihui Sun.
Validation: Xinwei Wang.
Visualization: Xinwei Wang.
Writing—original draft: Ye Li, Huihui Sun, Jie Guan, Tingting Ji, Xinwei Wang.
Writing—review & editing: Ye Li, Huihui Sun, Jie Guan, Tingting Ji, Xinwei Wang.
References
1. Piazuelo MB, Correa P. Gastric cancer: overview. Colomb Med (Cali). 2013; 44:192–201. PMID: 24892619.
2. Britto AV. [Stomach cancer: risk factors]. Cad Saude Publica. 1997; 13(Suppl 1):7–13.
3. Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol. 2009; 472:467–477. PMID: 19107449.

4. Choi NK, Youn KE, Heo DS, Lee SM, Kim Y, Park BJ. Stomach cancer incidence, mortality and survival rate in Korean Elderly Pharmacoepidemiologic Cohort (KEPEC) in 1994~1998. Cancer Res Treat. 2003; 35:383–390. PMID: 26680963.

5. Yoshida K, Yamaguchi K, Okumura N, Osada S, Takahashi T, Tanaka Y, et al. The roles of surgical oncologists in the new era: minimally invasive surgery for early gastric cancer and adjuvant surgery for metastatic gastric cancer. Pathobiology. 2011; 78:343–352. PMID: 22104206.
6. Layke JC, Lopez PP. Gastric cancer: diagnosis and treatment options. Am Fam Physician. 2004; 69:1133–1140. PMID: 15023013.
7. Lee EY, Lee YY, Suh M, Choi E, Mai TTX, Cho H, et al. Socioeconomic inequalities in stomach cancer screening in Korea, 2005–2015: after the introduction of the National Cancer Screening Program. Yonsei Med J. 2018; 59:923–929. PMID: 30187698.

8. McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014; 11:664–674. PMID: 25134511.

9. Zhang T, Liu C, Huang S, Ma Y, Fang J, Chen Y. A downmodulated microRNA profiling in patients with gastric cancer. Gastroenterol Res Pract. 2017; 2017:1526981. PMID: 28546810.

10. Ishimoto T, Baba H, Izumi D, Sugihara H, Kurashige J, Iwatsuki M, et al. Current perspectives toward the identification of key players in gastric cancer microRNA dysregulation. Int J Cancer. 2016; 138:1337–1349. PMID: 26041092.

11. Zhou S, Ye W, Ren J, Shao Q, Qi Y, Liang J, et al. MicroRNA-381 increases radiosensitivity in esophageal squamous cell carcinoma. Am J Cancer Res. 2014; 5:267–277. PMID: 25628936.
12. Liang Y, Zhao Q, Fan L, Zhang Z, Tan B, Liu Y, et al. Down-regulation of MicroRNA-381 promotes cell proliferation and invasion in colon cancer through up-regulation of LRH-1. Biomed Pharmacother. 2015; 75:137–141. PMID: 26320367.

13. Cao Q, Liu F, Ji K, Liu N, He Y, Zhang W, et al. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017; 36:29. PMID: 28193228.

14. Zhang M, Huang S, Long D. MiR-381 inhibits migration and invasion in human gastric carcinoma through downregulatedting SOX4. Oncol Lett. 2017; 14:3760–3766. PMID: 28927144.

15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. PMID: 11846609.
16. Zhou H, Yang Y, Li C, Yu B, Zhang S. Enhanced iridium complex electrochemiluminescence cytosensing and dynamic evaluation of cell-surface carbohydrate expression. Chemistry. 2014; 20:14736–14743. PMID: 25234357.

17. Bland JM, Altman DG. Survival probabilities (the Kaplan-Meier method). BMJ. 1998; 317:1572. PMID: 9836663.
18. Alberti C, Timsit JF, Chevret S. [Survival analysis - the log rank test]. Rev Mal Respir. 2005; 22(5 Pt 1):829–832. PMID: 16272989.
20. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016; 388:2654–2664. PMID: 27156933.

21. Wang J, Wu S, Huang T. Expression and role of VEGFA and miR-381 in portal vein tumor thrombi in patients with hepatocellular carcinoma. Exp Ther Med. 2018; 15:5450–5456. PMID: 29904424.

22. Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro' E, Levine AJ, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014; 33:5173–5182. PMID: 24166498.

23. Rothschild SI, Tschan MP, Jaggi R, Fey MF, Gugger M, Gautschi O. MicroRNA-381 represses ID1 and is deregulated in lung adenocarcinoma. J Thorac Oncol. 2012; 7:1069–1077. PMID: 22592211.

24. Li Y, Zhao C, Yu Z, Chen J, She X, Li P, et al. Low expression of miR-381 is a favorite prognosis factor and enhances the chemosensitivity of osteosarcoma. Oncotarget. 2016; 7:68585–68596. PMID: 27612424.

25. Chen Y, Min L, Zhang X, Hu S, Wang B, Liu W, et al. Decreased miRNA-148a is associated with lymph node metastasis and poor clinical outcomes and functions as a suppressor of tumor metastasis in non-small cell lung cancer. Oncol Rep. 2013; 30:1832–1840. PMID: 23843100.

26. Fujino Y, Takeishi S, Nishida K, Okamoto K, Muguruma N, Kimura T, et al. Downregulation of microRNA-100/microRNA-125b is associated with lymph node metastasis in early colorectal cancer with submucosal invasion. Cancer Sci. 2017; 108:390–397. PMID: 28032929.

27. Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014; 17:26–33. PMID: 23572188.

28. Ucar E, Semerci E, Ustun H, Yetim T, Huzmeli C, Gullu M. Prognostic value of preoperative CEA, CA 19-9, CA 72-4, and AFP levels in gastric cancer. Adv Ther. 2008; 25:1075–1084. PMID: 18821070.

29. Takahashi Y, Takeuchi T, Sakamoto J, Touge T, Mai M, Ohkura H, et al. The usefulness of CEA and/or CA19-9 in monitoring for recurrence in gastric cancer patients: a prospective clinical study. Gastric Cancer. 2003; 6:142–145. PMID: 14520526.

30. Mattar R, Alves de, DiFavero GM, Gama-Rodrigues JJ, Laudanna AA. Preoperative serum levels of CA 72-4, CEA, CA 19-9, and alpha-fetoprotein in patients with gastric cancer. Rev Hosp Clin Fac Med Sao Paulo. 2002; 57:89–92. PMID: 12118264.

31. Henry JC, Bassi C, Giovinazzo F, Bloomston M. MicroRNA from pancreatic duct aspirate differentiates cystic lesions of the pancreas. Ann Surg Oncol. 2013; 20(Suppl 3):S661–S666. PMID: 23884752.

32. Slattery ML, Herrick JS, Mullany LE, Samowitz WS, Sevens JR, Sakoda L, et al. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer. 2017; 56:769–787. PMID: 28675510.

33. Zhao FL, Dou YC, Wang XF, Han DC, Lv ZG, Ge SL, et al. Serum microRNA-195 is down-regulated in breast cancer: a potential marker for the diagnosis of breast cancer. Mol Biol Rep. 2014; 41:5913–5922. PMID: 25103018.

34. Zhang X, Nie Y, Li X, Wu G, Huang Q, Cao J, et al. MicroRNA-181a functions as an oncomir in gastric cancer by targeting the tumour suppressor gene ATM. Pathol Oncol Res. 2014; 20:381–389. PMID: 24531888.

35. Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, et al. Genomewide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012; 316:196–203. PMID: 22169097.

Fig. 1
The expression of serum miRNA-381, CA199, CA724, and CEA among healthy people, EGC patients, and AGC patients. (A) The expression of miRNA-381 in three groups. (B) The expression of CA199 in three groups. (C) The expression of CA724 in three groups. (D) The expression of CEA in three groups. One-way ANOVA was used; Tukey's multiple comparisons test was used to compare the two comparisons after ANOVA analysis. *Compared with healthy group, p<0.05, †compared with EGC group, p<0.05. CA, carbohydrate antigen; CEA, carcinoembryonic antigen; EGC, early gastric cancer; AGC, advanced gastric cancer.
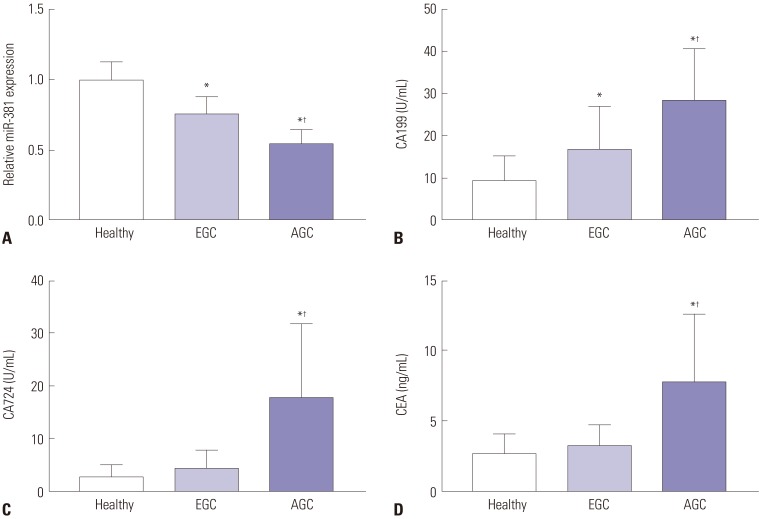
Fig. 2
AUC for miRNA-381, CA199, CA724, and CEA in gastric cancer patients and healthy individual. (A) The AUC values of miRNA-381, CA199, CA724, and CEA in EGC vs. healthy. (B) The AUC values of miRNA-381, CA199, CA724, and CEA in EGC vs. AGC. p<0.05 was considered a significant difference. AUC, area under the receiver operating characteristic curve; CA, carbohydrate antigen; CEA, carcinoembryonic antigen; EGC, early gastric cancer; AGC, advanced gastric cancer.
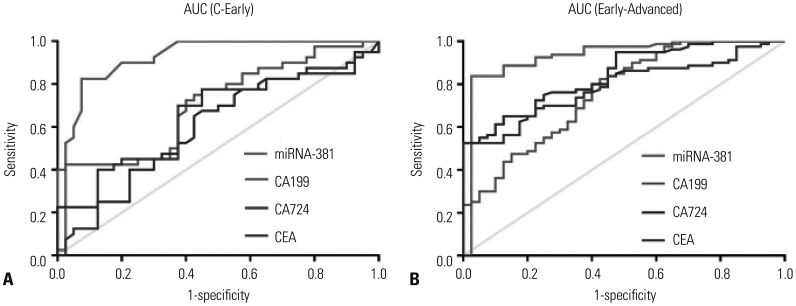
Fig. 3
Kaplan-Meier survival analysis for gastric cancer patients based on differential expression of miRNA-381. p<0.05 was considered as a significant difference.
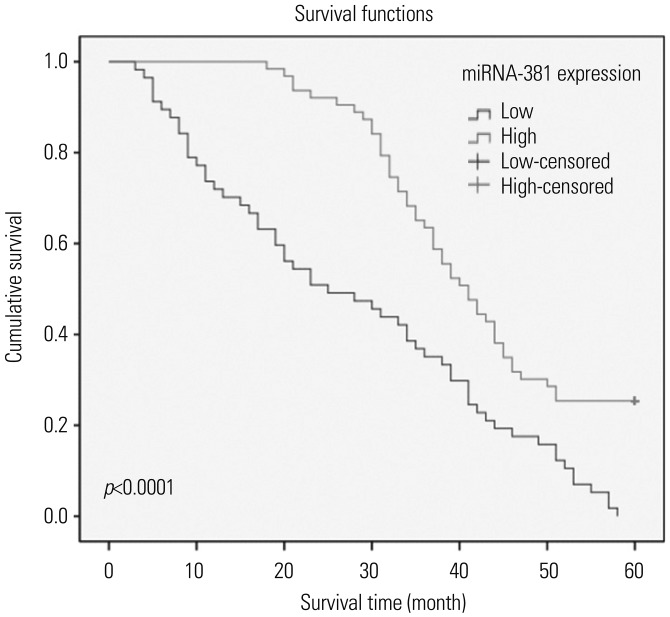
Table 1
Baseline Information for Study Participants
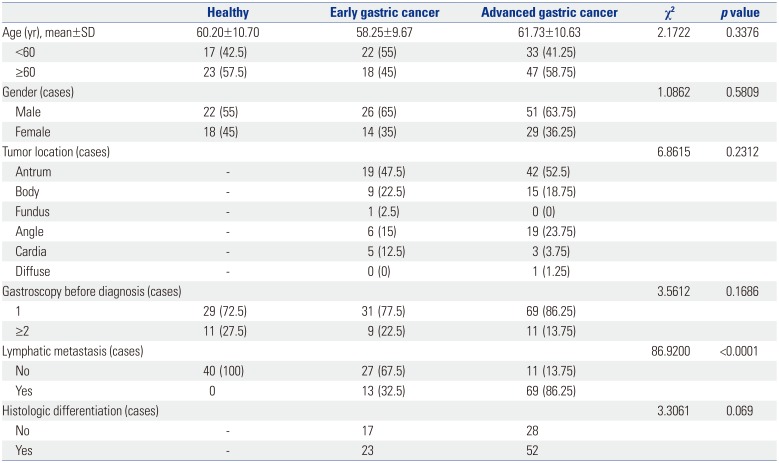
Table 2
Evaluation of Tumor Biomarkers
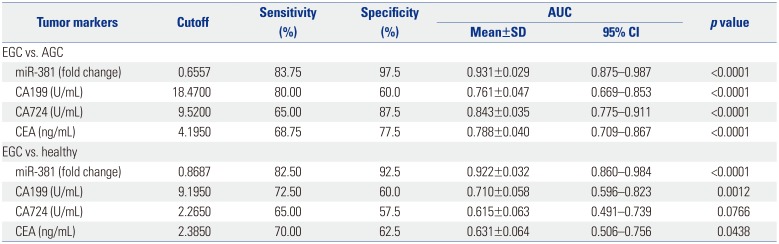
Table 3
Correlations between Expression of miRNA-381 and Clinicopathological Features




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download