1. Wheeler HM, Mlynash M, Inoue M, Tipirneni A, Liggins J, Zaharchuk G, et al. DEFUSE 2 Investigators. Early diffusion-weighted imaging and perfusion-weighted imaging lesion volumes forecast final infarct size in DEFUSE 2. Stroke. 2013; 44:681–685. PMID:
23390119.

2. Muir KW, Buchan A, von Kummer R, Rother J, Baron JC. Imaging of acute stroke. Lancet Neurol. 2006; 5:755–768. PMID:
16914404.

3. Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage Clin. 2017; 15:633–643. PMID:
28664034.

4. Mah YH, Jager R, Kennard C, Husain M, Nachev P. A new method for automated high-dimensional lesion segmentation evaluated in vascular injury and applied to the human occipital lobe. Cortex. 2014; 56:51–63. PMID:
23347558.

5. Boldsen JK, Engedal TS, Pedraza S, Cho TH, Thomalla G, Nighoghossian N, et al. Better diffusion segmentation in acute ischemic stroke through automatic tree learning anomaly segmentation. Front Neuroinform. 2018; 12:21. PMID:
29910721.

6. Straka M, Albers GW, Bammer R. Real-time diffusion-perfusion mismatch analysis in acute stroke. J Magn Reson Imaging. 2010; 32:1024–1037. PMID:
21031505.

7. Forbes F, Doyle S, Garcia-Lorenzo D, Barillot C, Dojat M. Adaptive weighted fusion of multiple MR sequences for brain lesion segmentation. In : 2010 IEEE international symposium on biomedical imaging: from nano to macro; 2010 April 14–17; Rotterdam, Netherlands.
8. Charoensuk W, Covavisaruch N, Lerdlum S, Likitjaroen Y. Acute stroke brain infarct segmentation in DWI images. Int J Pharm Med Biol Sci. 2015; 4:115–122.
9. Prakash KNB, Gupta V, Bilello M, Beauchamp NJ, Nowinski WL. Identification, segmentation, and image property study of acute infarcts in diffusion-weighted images by using a probabilistic neural network and adaptive Gaussian mixture model. Acad Radiol. 2006; 13:1474–1484. PMID:
17138115.
10. Mohd Saad N, Noor NSM, Abdullah AR, Muda S, Muda AF, Abdul Rahman NNS. Automated stroke lesion detection and diagnosis system. In : International MultiConference of engineers and computer scientists; 2017 March 15–17; Hong Kong, China.
11. Maier O, Wilms M, von der Gablentz J, Krämer UM, Münte TF, Handels H. Extra tree forests for sub-acute ischemic stroke lesion segmentation in MR sequences. J Neurosci Methods. 2015; 240:89–100. PMID:
25448384.

12. Peng Y, Zhang X, Hu Q. Segmentation of hyper-acute ischemic infarcts from diffusion weighted imaging based on support vector machine. Journal of Computer and Communications. 2015; 3:152–157.

13. Nag MK, Koley S, China D, Sadhu AK, Balaji R, Ghosh S, et al. Computer-assisted delineation of cerebral infarct from diffusion-weighted MRI using Gaussian mixture model. Int J Comput Assist Radiol Surg. 2017; 12:539–552. PMID:
28070776.

14. Warach S, Chien D, Li W, Ronthal M, Edelman RR. Fast magnetic resonance diffusion-weighted imaging of acute human stroke. Neurology. 1992; 42:1717–1723. PMID:
1513459.

15. van Everdingen KJ, van der Grond J, Kappelle LJ, Ramos LMP, Mali WPTM. Diffusion-weighted magnetic resonance imaging in acute stroke. Stroke. 1998; 29:1783–1790. PMID:
9731595.

16. Purushotham A, Campbell BC, Straka M, Mlynash M, Olivot JM, Bammer R, et al. Apparent diffusion coefficient threshold for delineation of ischemic core. Int J Stroke. 2015; 10:348–353. PMID:
23802548.

17. Ogata T, Christensen S, Nagakane Y, Ma H, Campbell BC, Churilov L, et al. EPITHET and DEFUSE Investigators. The effects of alteplase 3 to 6 hours after stroke in the EPITHET-DEFUSE combined dataset: post hoc case-control study. Stroke. 2013; 44:87–89. PMID:
23250996.
18. Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017; 18:570–584. PMID:
28670152.

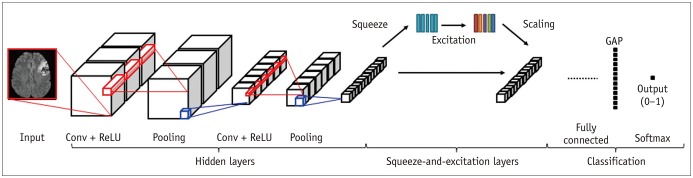
19. Woo I, Lee AR, Lee H, Choi K, Kang DW, Jung SC, et al. Sementic segmentation with squeeze-and-excitation block: application to infarct segmentation in DWI. In : NIPS 2017: the thirty-first annual conference on Neural Information Processing Systems; 2017 December 4–9; Long Beach, CA, USA.
20. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In : 18th Medical Image Computing and Computer Assisted Interventions (MICCAI); 2015 October 5–9; Munich, Germany.
21. Jégou S, Drozdzal M, Vazquez D, Romero A, Bengio Y. The one hundred layers tiramisu: fully convolutional DenseNets for semantic segmentation. In : IEEE Conference on Computer Vision and Pattern Recognition Workshop (CVPRW); 2017 July 21–26; Honolulu, HI, USA.
22. Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In : IEEE conference on Computer Vision and Pattern Recognition(CVPR); 2018 June 18–22; Salt Lake City, UT, USA.
23. Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. In : 27th international conference on international conference on machine learning; 2010 June 21–24; Haifa, Israel.
24. Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA, et al. Australian Imaging Biomarkers Lifestyle Flagship Study of Ageing. Alzheimer's Disease Neuroimaging Initiative. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014; 6:9–19. PMID:
25379412.

25. Noh H, Hong S, Han B. Learning deconvolution network for semantic segmentation. In : IEEE international conference on computer vision (ICCV); 2015 December 7–13; Santiago, Chile.
26. Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. In : IEEE conference on Computer Vision and Pattern Recognition(CVPR); 2017 July 21–26; Honolulu, HI, USA.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download