Dear Editor,
ABO is clinically the most important blood group system. The common O alleles of the ABO gene share a deletion of G at position 261 in exon 6, which induces a premature stop codon known to be responsible for inactivation of the glycosyltransferase properties [1]. Nondeletional O alleles constitute a major impediment in ABO genotyping based on targeted single nucleotide variation (SNV) analysis [2]. We describe a rare case of a nondeletional O allele, ABO*O.09.01, which has not been reported in Korea. Reporting of the present case without obtaining informed consent from the patient/parents was approved by the Institutional Review Board of Seoul National University Hospital, Seoul, Korea (H-1903-022-1015).
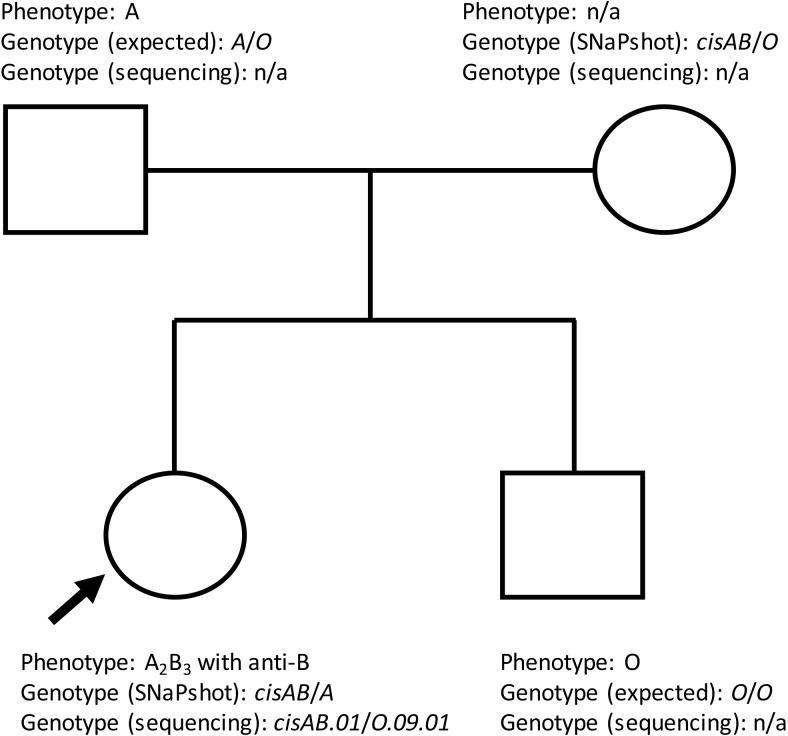
A 10-year-old girl visited Seoul National University Hospital in April 2018 with a one-year history of progressive hearing loss. She was diagnosed as having otosclerosis and was scheduled for laser stapedotomy. During the routine preoperative workup, her mother (whose genotype was previously confirmed to be cisAB/O by SNaPshot assay at our hospital) requested a consultation regarding the patient's ABO group.
The patient's ABO phenotype was serologically determined to be A2B3 with anti-B. The direct antiglobulin test (Bio-Rad, Hercules, CA, USA) was negative (anti-IgG+C3d), and the antibody screening (Bio-Rad) was negative in both panel cells. The ABO group of the patient's father and brother was additionally tested and was serologically typed as A and O, respectively (Fig. 1). Based on the information obtained from the family members, the genotypes were expected to be A/O (father), cisAB/O (mother), cisAB/O (patient), and O/O (brother). Molecular analysis was conducted for further evaluation.
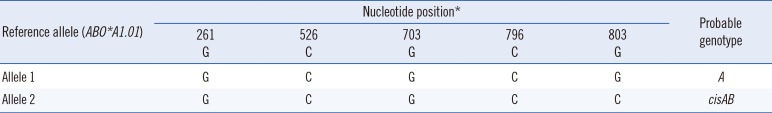
The ABO genotype was first determined by in-house allele-specific PCR using the SNaPshot assay (Applied Biosystems, Foster City, CA, USA) that was designed to identify SNVs at five nucleotide differentiation sites [3]. Briefly, genomic DNA was extracted by Gentra Puregene DNA Isolation Kit (Gentra Systems, Inc., Minneapolis, MN, USA) from peripheral blood, according to the manufacturer's instructions. PCR amplification using in-house allele-specific primer was done by the following conditions: 94℃ for 3 minutes followed by 30 cycles of 94℃ for 45 seconds, 68℃ for 2 minutes 30 seconds, and final extension at 72℃ for 5 minutes. Nucleotides 261, 526, 703, 796, and 803 in exons 6 and 7 of the ABO gene were analyzed using the Multiplex SNaPshot assay; this assay is used as a screening tool mainly to differentiate between common ABO alleles, especially when a patient is suspected to have a cisAB allele, rather than a variant in the A or B alleles. However, the SNaPshot assay showed that the patient's probable genotype was cisAB/A (Table 1). As the A1 antigen was not detected in her serological assay, it was clinically suspected that either the A allele expressed A2 or a weaker phenotype, or that a nondeletional O allele lacking c.261delG might be responsible for her A2 phenotype. For further investigation, we performed direct sequencing of exons 6 and 7 and the flanking regions, using in-house primers. Heterozygous base substitutions were detected at c.375–42A>G and 375–40G>A in intron 6 and c.467C>T, c.646T>A, c.681G>A, c.771C>T, c.803G>C, and c.829G>A in exon 7, corresponding to ABO* O.09.01 and ABO*cisAB01.
Nondeletional O alleles have been predominantly reported in Caucasians [2]. Determining O alleles merely by targeting c.261delG, as in the SNaPshot assay, may lead to misidentification of nondeletional O alleles as A alleles. However, as these genotypes are extremely rare in Asian population [4], less consideration was given to this possibility.
The phenotypes of individuals with cisAB vary widely [5]. Although our patient expressed A2B3 with anti-B, which is typical for cisAB/O, variant ability to synthesize the A antigen has been reported for nondeletional O alleles [67]. Ogasawara, et al. [8] first reported ABO*O.09.01 (previously known as R102) and noted a difference in the weak reactivity with monoclonal anti-A between R102/B and non-R102/B family members. This serological difference was explained by allelic enhancement [9]. Hosseini-Maaf, et al. [10] argued that nondeletional O alleles may cause weak A expression on red blood cells because of autologous chimerism. They hypothesized that O alleles with different defects may exchange genetic material between alleles to produce functional A alleles by recombination or conversion. ABO* AW.31.02, ABO*AW.31.03, ABO*AW.31.04, ABO*AW.31.05, and ABO*O.09.01 all have identical coding sequences but different intron 6 sequences, which may be related to their resulting phenotype of either weak A or O. Unfortunately, we did not perform adsorption/elution assays to detect the expression of weak ABO antigens in the patient's brother and father.
In conclusion, this case highlights the importance of comparing the serological phenotype and family history when interpreting ABO genotyping assays based on targeted SNV analysis as it may lead to misleading results. The absence of c.261delG in O alleles should be also considered in the Korean population.
Notes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
Author Contributions: Dahae Yang wrote the manuscript; Boram Kim, Da Young Song, Man Jin Kim, and Sung Im Cho conducted laboratory work and also described the process and interpreted the results; and Tae Yeul Kim, Hyungsuk Kim, Moon-Woo Seong, and Sung Sup Park contributed to the writing and revision of the manuscript.
References
1. Yip SP. Sequence variation at the human ABO locus. Ann Hum Genet. 2002; 66:1–27. PMID: 12014997.
2. Wagner FF, Blasczyk R, Seltsam A. Nondeletional ABO*O alleles frequently cause blood donor typing problems. Transfusion. 2005; 45:1331–1334. PMID: 16078922.
3. Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990; 345:229–233. PMID: 2333095.
4. Song SH, Chang HE, Ryu KC, Lee HJ, Park KU, Song J, et al. Analysis for eight ABO alleles in Korean population. Korean J Lab Med. 2006; 26:374–379. PMID: 18156754.
5. Park MS, Chun S, Lee CH, Cho D. Diverse phenotypes of cis-AB blood group and transfusion strategy. Korean J Blood Transfus. 2016; 27:304–306.
6. Yazer MH, Hosseini-Maaf B, Olsson ML. Blood grouping discrepancies between ABO genotype and phenotype caused by O alleles. Curr Opin Hematol. 2008; 15:618–624. PMID: 18832934.
7. Seltsam A, Das Gupta C, Wagner FF, Blasczyk R. Nondeletional ABO*O alleles express weak blood group A phenotypes. Transfusion. 2005; 45:359–365. PMID: 15752153.
8. Ogasawara K, Yabe R, Uchikawa M, Nakata K, Watanabe J, Takahashi Y, et al. Recombination and gene conversion-like events may contribute to ABO gene diversity causing various phenotypes. Immunogenetics. 2001; 53:190–199. PMID: 11398963.
9. Olsson ML, Michalewska B, Hellberg A, Walaszczyk A, Chester MA. A clue to the basis of allelic enhancement: occurrence of the Ax subgroup in the offspring of blood group O parents. Transfus Med. 2005; 15:435–442. PMID: 16202060.
10. Hosseini-Maaf B, Irshaid NM, Hellberg A, Wagner T, Levene C, Hustinx H, et al. New and unusual O alleles at the ABO locus are implicated in unexpected blood group phenotypes. Transfusion. 2005; 45:70–81. PMID: 15647021.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download