This article has been
cited by other articles in ScienceCentral.
Dear Editor,
Mycobacterium virginiense is a recently described species of the
Mycobacterium terrae complex (MTC) [
12]. It is recognized as an infectious agent of clinical importance [
3].
M. virginiense is a slow-growing nontuberculous mycobacterium (NTM) that was first identified in 2016 along with three other validated clinical strains causing tenosynovitis and osteomyelitis [
1]. The three
M. virginiense isolates were nonchromogenic on Middlebrook 7H10 agar (Sigma-Aldrich, St. Louis, MO, USA) and were resistant to several antibiotics, including rifampin and quinolones.
M. virginiense had never been isolated from the human body until Vasireddy, et al. [
12] reported three isolates from the tendon, elbow, and knee. Since the first report in 2016, only two more
M. virginiense isolates, one from a mud specimen of a swine farm in Japan [
4] and another from bovine fecal specimens [
5], have been reported. We report the first isolation of
M. virginiense from a human pulmonary specimen. This study was approved by the Institutional Review Board of Samsung Medical Center, Seoul, Korea (No. 2019-05-117), which waived the need for informed consent from the patient.
A 67-year-old male patient visited the pulmonary department every year at Samsung Medical Center because of NTM infection. He had a history of tuberculous pleurisy at age 21 and had been diagnosed as having NTM pulmonary disease caused by Mycobacterium massiliense at age 54. He had been treated for two yrs with an antibiotic regimen that included clarithromycin and ciprofloxacin until his sputum culture showed negative results. He had been diagnosed as having chronic pulmonary aspergillosis at age 62. The patient submitted sputum specimens every year for acid-fast bacilli staining and culture testing. NTM has been repeatedly isolated from three yearly consecutive sputum specimens since the age of 65.
Sputum cultures were performed using liquid media with Middlebrook 7H9 broth in an MGIT 960 system (Becton Dickinson, Sparks, MD, USA) and using solid media with 3% Ogawa agar (Shinyang, Seoul, Korea). A line probe assay for the internal transcribed spacer gene (AdvanSure Mycobacteria GenoBlot Assay; LG Chem, Seoul, Korea) revealed that all three isolates were unidentifiable
Mycobacterium species. For definitive species identification, 16S rDNA and
rpoB genes from two out of three isolates were sequenced according to the protocol outlined in the Clinical and Laboratory Standards Institute (CLSI) guidelines MM18-A [
6]. Using the basic local alignment search tool (BLAST) algorithm, we found that the 16S rDNA and
rpoB sequences exhibited 100% (518/518 bp) and 99.4% (340/342 bp) similarity, respectively, to those of
M. virginiense. The next closest matches were
Mycobacterium paraterrae at 99.6% (516/518 bp) for 16S rRNA sequences and
Mycobacterium sinense at 95.2% (334/351 bp) for
rpoB sequences. The organism was finally identified as
M. virginiense for both specimens. Phylogenetic trees with a bootstrapping value of 1,000 were constructed based on the 16S rRNA and
rpoB sequences using MEGA-X software (
https://www.megasoftware.net;
Fig. 1). Antimicrobial susceptibility was tested at the Korean Institute of Tuberculosis using the broth microdilution method as described in the CLSI guidelines M24-A2 (
Table 1) [
7]. A computed tomography (CT) scan of the chest revealed that the left lung was damaged by tuberculosis and NTM disease sequela.
The patient's lung had been totally destroyed by
M. massiliense infection in 2006, and the CT scan revealed neither improvement nor deterioration ever since
M. virginiense was detected. The patient's symptoms had not changed in the past five years. Thus, there is insufficient evidence to conclude that his NTM lung disease was caused by
M. virginiense pathogenicity. While
M. virginiense is recognized as a primary cause of tenosynovitis and osteomyelitis [
18], whether this species is a true respiratory pathogen has not been established. However, the positive culture results in three consecutive years in the present study may indicate a link between
M. virginiense and pulmonary disease. Of note, the patient works for the Korean Forest Service; thus, he is frequently exposed to mud and soil. Because
M. virginiense has been isolated from mud and bovine fecal specimens, its infection in this case may be linked to the patient's professional activities [
45].
Because of insufficient clinical data, there is no standard treatment for
M. virginiense infection. The strain from our case showed a high minimum inhibitory concentration (MIC) to rifampicin and the quinolones (ciprofloxacin and moxifloxacin). Previous studies have indicated the therapeutic benefits of a macrolide, combined with additional agents, to treat MTC tenosynovitis [
18]. Before antimicrobial selection strategies can be developed, more research is needed on
M. virginiense's pathogenicity, association with NTM lung disease, and antimicrobial susceptibility.
Figures and Tables
 | Fig. 1Neighbor-joining phylogenetic trees for Mycobacterium virginiense and other Mycobacterium species based on (A) the 16S rRNA gene and (B) the rpoB gene. GenBank accession numbers are given in parentheses.
|
Table 1
Antimicrobial susceptibility of Mycobacterium virginiense
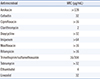
|
Antimicrobial |
MIC (μg/mL) |
|
Amikacin |
> 128 |
|
Cefoxitin |
32 |
|
Ciprofloxacin |
> 16 |
|
Clarithromycin |
2 |
|
Doxycycline |
> 32 |
|
Imipenem |
> 64 |
|
Moxifloxacin |
> 16 |
|
Rifampicin |
> 16 |
|
Trimethoprim/sulfamethoxazole |
16/304 |
|
Tobramycin |
> 32 |
|
Ethambutol |
4 |
|
Linezolid |
32 |








