Dear Editor,
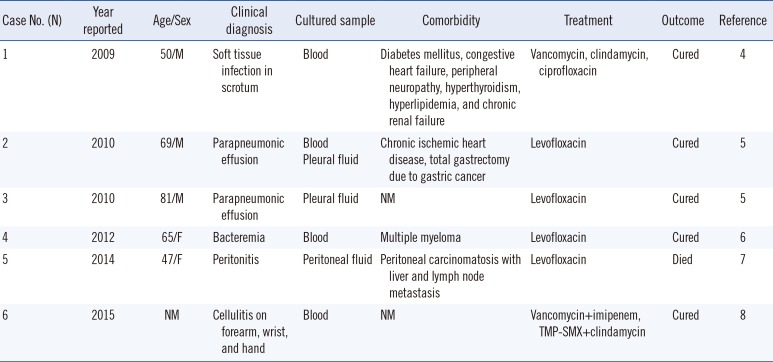
Aureimonas altamirensis, a gram-negative, nonmotile bacterium that grows in strictly aerobic environments, was isolated from the subterranean environment of Altamira Cave in Spain in 2006 and was designated Aurantimonas altamirensis [1]; however, it was reclassified as Aureimonas altamirensis with other Aurantimonas species. In 2008, it was first isolated from clinical samples [2]. A. altamirensis has usually been implicated as a contaminant originating from the environment, such as water sources [2], and is reported as an agent of canine infection [3]. To date, only a few cases of human infections have been reported (Table 1) [45678]. We report the first case of bacteremia by A. altamirensis in Korea. The Institutional Review Board of Chonbuk National University Hospital, Jeonju, Korea, determined that this study has a low ethical load and did not require informed consent from the patient (IRB No. CUH 2018-03-028).
A 65-year-old male presented to the emergency room of Chonbuk National University Hospital because of fever and general weakness in January 2015. Five and 10 years previously, he had been diagnosed as having diabetes mellitus and a stroke, respectively, but had been well prior to admission. On initial physical examination, the patient exhibited no unusual features, and his vital signs were normal except for a body temperature of 38.9℃. His white blood cell count was 19×109/L (neutrophils 96%), Hb level was 12.3 g/L, and platelet count was 380×109/L. Total bilirubin and direct bilirubin levels were elevated at 49.26 and 43.27 µmol/L, respectively. His AST level was 0.99 µkat/L, ALT level was 1.00 µkat/L, glycated Hb (HbA1c) level was 6.5%, and C-reactive protein level was 1,971.47 nmol/L in initial blood tests. Abdominal computed tomography showed unremarkable findings. Two sets of blood culture (FA Plus, FN Plus, BacT/Alert 3D system, bioMérieux, Durham, NC, USA) were done aseptically, and one of them was positive. From the subculture on blood agar plate incubated on 35℃ with non-CO2 condition for 24 hours, grayish colonies were observed. They were gram-negative bacilli that were catalase- and oxidase-positive. The isolate did not grow well in a 5% CO2 condition; it grew well on both blood agar and MacConkey agar plates under non-CO2 conditions but did not produce any colonies on MacConkey agar plates when incubated with 5% or 10% CO2 for 24 hours. The Vitek 2 system (bioMérieux Inc., Hazelwood, MO, USA) identified this organism as Acinetobacter lwoffii (96% probability), while the Vitek MS system (bioMérieux, Marcy L'Étoile, France) could not identify the organism. 16S ribosomal RNA sequencing (GenBank, MK749933) showed 99.6% (1,361/1,366) homology with Aureimonas altamirensis (GenBank, NR_043764.1), and the % difference with the second most highly matched organism, Aureimonas frigidaquae (GenBank, NR_044195.1), was 98.4% (1,357/1,379). A phylogenetic tree analysis with representative Aurantimonas strains downloaded from BLAST (http://www.ncbi.nlm.nih.gov/blast) was performed using MEGA X (https://www.megasoftware.net) based on the 16S ribosomal RNA sequences [59]. The results showed that our isolate formed a clade with the other A. altamirensis strains (Fig. 1).
Intravenous moxifloxacin was empirically administered, and the patient's condition improved. We performed positron emission tomography-computed tomography with [18F]-fluorodeoxyglucose to investigate the possible causes of fever. However, the results were normal, and repeated physical examinations and imaging studies showed that the patient did not have any other infection. In addition, the follow-up blood cultures were all negative, and the patient showed a good clinical response to empirical antibiotics. Therefore, we believed that the patient had primary A. altamirensis bacteremia according to the clinical information obtained from 16S ribosomal RNA sequencing. The fever subsided approximately two weeks after antibiotic treatment, and the patient was discharged.
We performed an antimicrobial susceptibility testing using the E-test (bioMérieux, Marcy L'Étoile, France) after receiving the 16S ribosomal RNA sequencing result. The minimum inhibitory concentrations for amoxicillin/clavulanic acid, gentamicin, imipenem, and trimethoprim/sulfamethoxazole were 1/0.5, 2, 0.125, and >32/608 µg/mL, respectively. A. altamirensis was susceptible to amoxicillin/clavulanic acid and imipenem, according to the non-species related breakpoints specified by the European Committee on Antimicrobial Susceptibility Testing guidelines [10].
A PubMed Medline search identified six cases of invasive human infection by A. altamirensis from blood, pleural fluid, and peritoneal fluid. All patients clinically improved with antibiotics, and only one patient died because of underlying comorbidities [7].
To date, the clinical relevance of A. altamirensis remains uncertain [5]. However, based on previous reports and our study, it is difficult to negate its potential pathogenic significance in humans. Information on the susceptibility of A. altamirensis to most antibiotics is limited, and clinicians rely on only a few case reports and empirical treatments. However, most clinical isolates were susceptible to the major antibiotics used for gram-negative bacteria, except quinolone, and showed successful treatment responses [2457].
In conclusion, although there is limited information on human infections, A. altamirensis should be considered an opportunistic pathogen, especially in immunocompromised patients. Additionally, laboratory physicians should be careful when there are discrepancies between identification results from automated systems and biochemical tests with growth conditions.
Acknowledgment
This paper was supported by the Fund of Biomedical Research Institute, Chonbuk National University Hospital.
References
1. Jurado V, Gonzalez JM, Laiz L, Saiz-Jimenez C. Aurantimonas altamirensis sp. nov., a member of the order Rhizobiales isolated from Altamira Cave. Int J Syst Evol Microbiol. 2006; 56:2583–2585. PMID: 17082395.
2. Luong ML, Békal S, Vinh DC, Lauzon D, Leung V, Al-Rawahi GN, et al. First report of isolation and characterization of Aurantimonas altamirensis from clinical samples. J Clin Microbiol. 2008; 46:2435–2437. PMID: 18495852.
3. Reilly TJ, Calcutt MJ, Wennerdahl LA, Williams F 3rd, Evans TJ, Ganjam IK, et al. Isolation of Aureimonas altamirensis, a Brucella canis-like bacterium, from an edematous canine testicle. J Vet Diagn Invest. 2014; 26:795–798. PMID: 25292192.
4. Mendes RE, Denys GA, Fritsche TR, Jones RN. Case report of Aurantimonas altamirensis bloodstream infection. J Clin Microbiol. 2009; 47:514–515. PMID: 19036934.
5. Téllez-Castillo CJ, González Granda D, Bosch Alepuz M, Jurado Lobo V, Saiz-Jimenez C, Juan JL, et al. Isolation of Aurantimonas altamirensis from pleural effusions. J Med Microbiol. 2010; 59:1126–1129. PMID: 20558586.
6. García-Lozano T, Aznar Oroval E, Juan Bañón JL. First isolation in Spain of Aurantimonas altamirensis in a blood culture from a port-a-cath in a patient with Bence-Jones type multiple myeloma. Enferm Infecc Microbiol Clin. 2012; 30:217–218. PMID: 22361566.
7. Schröttner P, Rudolph WW, Taube F, Gunzer F. First report on the isolation of Aureimonas altamirensis from a patient with peritonitis. Int J Infect Dis. 2014; 29:71–73. PMID: 25461238.
8. Eshaghi A, Shahinas D, Patel SN, Kus JV. First draft genome sequence of Aureimonas altamirensis, isolated from patient blood culture. FEMS Microbiol Lett. 2015; 362.
9. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018; 35:1547–1549. PMID: 29722887.
10. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0. 2019. Updated on Jan 2019. http://www.eucast.org.
Fig. 1
Phylogenetic tree analysis based on 16S ribosomal RNA gene sequences. The tree was constructed using the neighbor-joining method with bootstrap resampling (1,000 replicates). The tree was rooted with Fluvimarina pelagi HTCC 2615 as the outgroup. Bootstrap values are expressed as percentages of 1,000 replications, and bars represent 0.010 substitutions per nucleotide position. GenBank database accession numbers are provided near the species names.
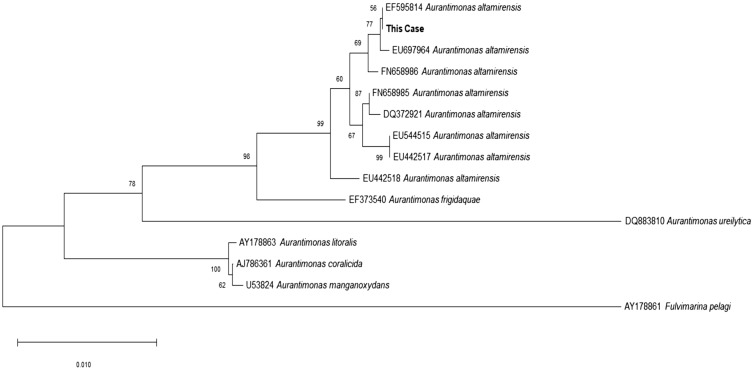
Table 1
Invasive Aureimonas altamirensis infections in humans




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download