Dear Editor,
Spinal muscular atrophy (SMA) is caused by homozygous deletion of the survival motor neuron gene (SMN1), and copy-number variation (CNV) in the SMN2 gene is thought to influence the disease severity.1 We present two late-onset SMA siblings who manifested with marked differences in clinical severity and muscle imaging despite the same copy numbers of SMN2.
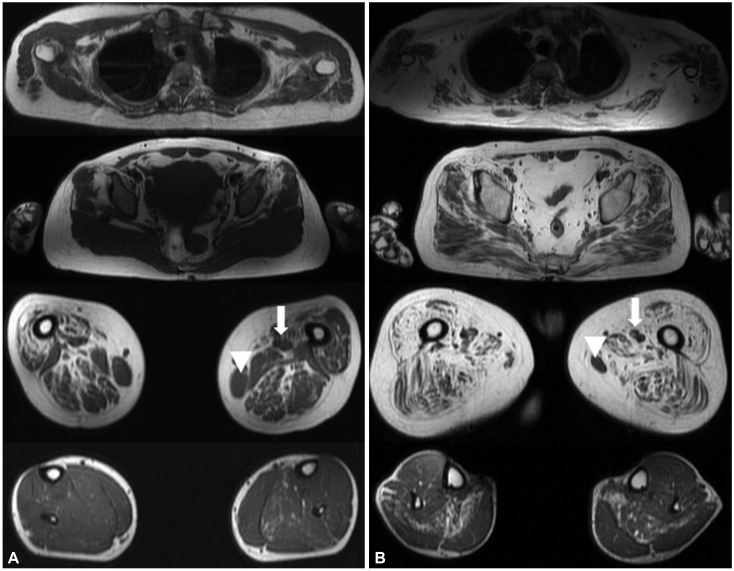
A 43-year-old female presented with proximal limb weakness since high school that had not deteriorated until she reached 40 years of age. The initial neurological examination of the proband demonstrated proximal limb weakness in the lower extremities, at MRC grade 4. However, she had no difficulty in performing the activities of daily living, but experienced difficulties during strenuous exercise such as hiking. The proband's elder brother was 45 years old, and he had found it difficult to run during high school, and had been incapable of independent walking since his 30s. The initial neurological examination revealed a proximal limb weakness with the lower extremity being affected more, and he was nonambulatory. Muscle MRI revealed that the proximal thigh muscles were markedly affected with relative sparing of the adductor and gracilis muscles (Fig. 1A). Her muscle biopsy showed fiber-type grouping, implicating neurogenic changes (Supplementary Fig. 1 in the online-only Data Supplement). The muscle MRI of the proband's elder brother illustrated a diffuse fatty change in proximal muscles including gluteus maximus, gluteus medius vastus, and hamstring muscles, but the gracilis and adductor longus muscles were mildly spared (Fig. 1B). A gene study of SMN1 revealed deletion of exons 7 and 8, confirming the diagnosis of SMA; the proband was classified as SMA4, while it was more appropriate to classify her brother as SMA3b. We measured the copy number of SMN2 of these two siblings using two different methods to ensure a precise diagnosis. We compared the SMN2, NAIP, and CFTR copy numbers as described previously,1 and we additionally used droplet digital PCR analysis (Bio-Rad Laboratories; ddPCR SMN2 copy number determination kit, Hercules, CA, USA). The obtained results were further confirmed by multicopy marker analysis as described by Melki et al.2 Both patients showed four copies of SMN2 and the same NAIP and CFTR copy numbers. Next-generation sequencing was also performed to screen the variants in DYNC1H1, BICD2, SMN1, PLS3, and NCALD, but this revealed no pathogenic mutation or known modifier known to affect the phenotype of our patients.
The CNVs of SMN2 are generally known to be correlated with the phenotype.1345 However, there are reports of phenotype discrepancies in patients with same copy numbers of SMN2.45 This means that variability in the phenotype cannot be reliably explained only by CNVs, and so we expect that there are other unknown genetic modifiers, sex differences, or epigenetic factors. It has reported that there are sex-related differences in severity or in intrafamilial phenotype variability.4678 A recent clinical study of myotonic dystrophy type 1 showed that sex differences might be another modifying factor influencing the clinical profile and severity of the disease.6 Consistent with these studies, we observed a significantly more severe phenotype in the male patient but a mild phenotype in the female patient of the same family. Therefore, even in SMA, sex difference is a possible modifying factor associated with phenotype variability, and further studies are warranted to clarify this.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (Ministry of Science and ICT) (Grant Nos. 2017R1C1B5076264 and NRF-2018R1C1B5045675).
References
1. Tran VK, Sasongko TH, Hong DD, Hoan NT, Dung VC, Lee MJ, et al. SMN2 and NAIP gene dosages in Vietnamese patients with spinal muscular atrophy. Pediatr Int. 2008; 50:346–351. PMID: 18533950.

2. Melki K, Lefebvre S, Burglen L, Burlet P, Clermont O, Millasseau P, et al. De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science. 1994; 264:1474–1477. PMID: 7910982.

3. Durmus H, Yilmaz R, Gulsen-Parman Y, Oflazer-Serdaroglu P, Cuttini M, Dursun M, et al. Muscle magnetic resonance imaging in spinal muscular atrophy type 3: selective and progressive involvement. Muscle Nerve. 2017; 55:651–656. PMID: 27543937.

4. Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time LightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002; 70:358–368. PMID: 11791208.

5. Wirth B, Herz M, Wetter A, Moskau S, Hahnen E, Rudnik-Schöneborn S, et al. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet. 1999; 64:1340–1356. PMID: 10205265.

6. Raymond K, Levasseur M, Mathieu J, Desrosiers J, Gagnon C. A 9-year follow-up study of the natural progression of upper limb performance in myotonic dystrophy type 1: a similar decline for phenotypes but not for gender. Neuromuscul Disord. 2017; 27:673–682. PMID: 28527585.

7. Jedrzejowska M, Milewski M, Zimowski J, Borkowska J, Kostera-Pruszczyk A, Sielska D, et al. Phenotype modifiers of spinal muscular atrophy: the number of SMN2 gene copies, deletion in the NAIP gene and probably gender influence the course of the disease. Acta Biochim Pol. 2009; 56:103–108. PMID: 19287802.

8. Cuscó I, Barceló MJ, Rojas-García R, Illa I, Gámez J, Cervera C, et al. SMN2 copy number predicts acute or chronic spinal muscular atrophy but does not account for intrafamilial variability in siblings. J Neurol. 2006; 253:21–25. PMID: 15981080.

Supplementary Materials
The online-only Data Supplement is available with this article at https://doi.org/10.3988/jcn.2019.15.3.395.
Supplementary Fig. 1
Histopathological findings in a biopsy of the deltoid muscle of the proband. The muscle fibers did not show significant necrosis or regenerating fibers, with well-organized intramyofibrillar networks (A, B). However, typical grouped fiber atrophy was observed, reflecting neurogenic changes (C, D, E). The dystrophin stain was normal (F) (A: Hematoxylin-eosin, ×200; B: NADH-tetrazolium reductase, ×100; C: Cytochrome c oxidase, ×100; D: ATPase 4.4, ×40; E: ATPase 4.4, ×100; and F: Dystrophin, ×200).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download