Abstract
Background
Blood culture is an important method for identifying infectious microorganisms and confirming that a selected antimicrobial treatment is appropriate. In this study, we investigated the annual changes in the frequencies of blood isolates and antibiotic susceptibility test (AST) results.
Methods
We created a large database comprising data on all patient-unique blood cultures obtained from January 2007 through December 2016. Blood specimens were cultured using the BD BACTEC FX system, and species identification and AST were performed using the VITEK 2 system.
Results
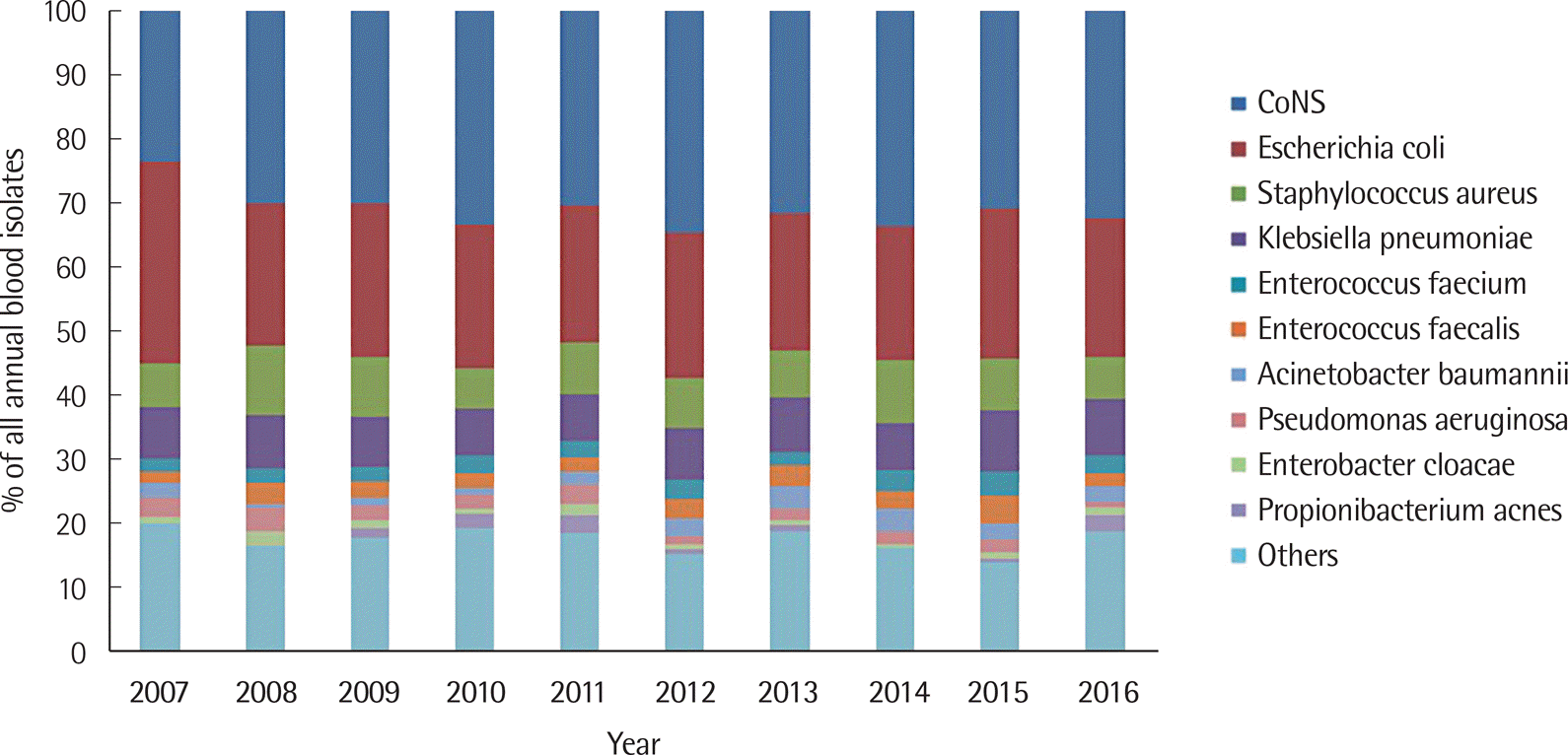
During the 10-year study period, a total of 203,651 blood culture results were collected. Of these, gram-positive cocci, gram-negative rods, and fungi were isolated in 2.15%, 0.55%, and 0.12% of the blood cultures, respectively. Escherichia coli was the most commonly isolated species (22.8%), followed by Staphylococcus epidermidis (16.8%), Klebsiella pneumoniae (8.1%), and Staphylococcus aureus (8.0%). Fungal species were isolated in 3.0% of all positive blood cultures. Candida albicans was the most commonly isolated species (1.1%), followed by Candida parapsilosis (0.6%). Methicillin resistance was seen in 55.2% of S. aureus isolates. The frequencies of vancomycin-resistant Enterococcus (VRE) and carbapenem-resistant Pseudomonas aeruginosa (CRPA) were 13.1% and 10.9%, respectively. The isolation rates of MRSA, VRE, and CRPA showed different patterns each year.
Go to : 
References
1. Weinstein MP, Towns ML, Quartey SM, Mirrett S, Reimer LG, Parmigiani G, et al. The clinical signifcance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis. 1997; 24:584–602.
2. Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD. The beneft of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med. 1998; 244:379–86.
3. Decousser JW, Lamy B, Pina P, Allouch PY. Trends in antibiotic susceptibility of bloodstream pathogens in hospitalized patients in France, 1996 to 2007. Diagn Microbiol Infect Dis. 2010; 66:292–300.

4. Biedenbach DJ, Moet GJ, Jones RN. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997–2002). Diagn Microbiol Infect Dis. 2004; 50:59–69.

5. Streit JM, Jones RN, Sader HS, Fritsche TR. Assessment of pathogen occurrences and resistance profles among infected patients in the intensive care unit: report from the SENTRY Antimicrobial Surveillance Program (North America, 2001). Int J Antimicrob Agents. 2004; 24:111–8.
6. Kang BK, Lee HJ, Suh JT. The trends of the species and antimicrobial susceptibility of bacteria and fungi isolated from blood cultures (1986–1996). Korean J Clin Pathol. 1998; 18:57–64.
7. Kim SY, Lim G, Kim MJ, Suh JT, Lee HJ. Trends in fve-year blood cultures of patients at a university hospital (2003∼2007). Korean J Clin Microbiol. 2009; 12:163–8.
8. Koh EM, Lee SG, Kim CK, Kim M, Yong D, Lee K, et al. Microorganisms isolated from blood cultures and their antimicrobial susceptibility patterns at a university hospital during 1994–2003. Korean J Lab Med. 2007; 27:265–75.

9. Clinical and Laboratory Standards Institute. Analysis and presentation of cumulative antimicrobial susceptibility test data; Approved guideline-Fourth edition. CLSI document M39-A4. Wayne PA:. Clinical and Laboratory Standards Institute;2014.
10. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 28th ed.CLSI supplement, M100. Wayne PA: Clinical and Laboratory Standards Institute;2018. p. 296.
11. Arpi M, Victor MA, M⊘ller JK, J⊘nsson V, Hansen MM, Peterslund NA, et al. Changing etiology of bacteremia in patients with hematological malignancies in Denmark. Scand J Infect Dis. 1994; 26:157–62.

12. Worth LJ and Slavin MA. Bloodstream infections in haematology: risks and new challenges for prevention. Blood Rev. 2009; 23:113–22.
13. Weinstein MP. Blood culture contamination: persisting problems and partial progress. J Clin Microbiol. 2003; 41:2275–8.

14. Favre B, Hugonnet S, Correa L, Sax H, Rohner P, Pittet D. Nosocomial bacteremia: clinical signifcance of a single blood culture positive for coagulase-negative staphylococci. Infect Control Hosp Epidemiol. 2005; 26:697–702.
15. Murni IK, Duke T, Daley AJ, Kinney S, Soenarto Y. True pathogen or contamination: validation of blood cultures for the diagnosis of nosocomial infections in a developing country. J Trop Pediatr. 2018; 64:389–94.

16. Suh JT, Chong YS, Park JW, Kim KD, Son HC, Yang DW, et al. A study on Salmonella species isolated from several university hospitals in Korea. J Korean Med Assoc. 1989; 32:1230–8.
17. Lee HJ. Serovars and antimicrobial susceptibility of the recent clinical isolates of Salmonella. Korean J Clin Pathol. 1995; 15:422–9.
18. Shin KS. Species and antimicrobial susceptibilities of bacteria isolated from blood during recent 5 years at Chungbuk National University Hospital. Chungbuk Med J. 2004; 14:61–9.
19. Vidal F, Mensa J, Almela M, Olona M, Martinez JA, Marco F, et al. Bacteraemia in adults due to glucose non-fermentative Gram-negative bacilli other than P. aeruginosa. QJM. 2003; 96:227–34.

20. Lai CH, Chi CY, Chen HP, Chen TL, Lai CJ, Fung CP, et al. Clinical characteristics and prognostic factors of patients with Stenotrophomonas maltophilia bacteremia. J Microbiol Immunol Infect. 2004; 37:350–8.
21. Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin infect Dis. 1999; 29:239–44.

22. Wey SB, Mori M, Pfaller MA, Woolson RF, Wenzel RP. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch Intern Med. 1988; 148:2642–5.

23. Kao AS, Brandt ME, Pruitt WR, Conn LA, Perkins BA, Stephens DS, et al. The epidemiology of candidemia in two United States cities: results of a population-based active surveillance. Clin Infect Dis. 1999; 29:1164–70.

24. Ji M, Choi YM, Bae E, Kim CK. Trends in microorganisms isolated from blood cultures at a veterans hospital from 2012 to 2015. J Lab Med Qual Assur. 2017; 39:141–6.

25. Yong D, Shin HB, Kim YK, Cho J, Lee WG, Ha GY, et al. Increase in the prevalence of carbapenem-resistant Acinetobacter isolates and ampicillin-resistant non-typhoidal Salmonella species in Korea: a KONSAR study conducted in 2011. Infect Chemother. 2014; 46:84–93.
26. Lee K, Lee MA, Lee CH, Lee J, Roh KH, Kim S, et al. Increase of ceftazidime- and fluoroquinolone-resistant Klebsiella pneumoniae and imipenem-resistant Acinetobacter spp. in Korea: analysis of KON-SAR study data from 2005 and 2007. Yonsei Med J. 2010; 51:901–11.
27. Kim SH, Lee KJ, Park C, Lee H. Results of Korean antimicrobial resistance monitoring system (KARMS) in general hospitals, 2008–2015. http://www.cdc.go.kr/CDC/info/CdcKrInfo0301.jsp?menuIds=. HOME006-MNU3003-MNU2950-MNU2951&cid=76882 (Updated on Nov 2017).
28. Gouby A, Neuwirth C, Bourg G, Bouziges N, Carles-Nurit MJ, Des-paux E, et al. Epidemiological study by pulsed-feld gel electrophoresis of an outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a geriatric hospital. J Clin Microbiol. 1994; 32:301–5.
29. Murray PR, Baron EJ, et al. eds. Manual of clinical microbiology. 7th ed.Washington: ASM Press;1999.
30. Farber BF, Eliopoulos GM, Ward JI, Ruoff K, Moellering RC Jr. Resistance to penicillin-streptomycin synergy among clinical isolates of viridans streptococci. Antimicrob Agents Chemother. 1983; 24:871–5.

31. Seo YH, Jeong JH, Lee HT, Kwoun WJ, Park PW, Ahn JY, et al. Analysis of blood culture data at a tertiary university hospital, 2006–2015. Ann Clin Microbiol. 2017; 20:35–41.

32. Rahkonen M, Luttinen S, Koskela M, Hautala T. True bacteremias caused by coagulase negative Staphylococcus are diffcult to distinguish from blood culture contaminants. Eur J Clin Microbiol Infect Dis. 2012; 31:2639–44.
Go to : 
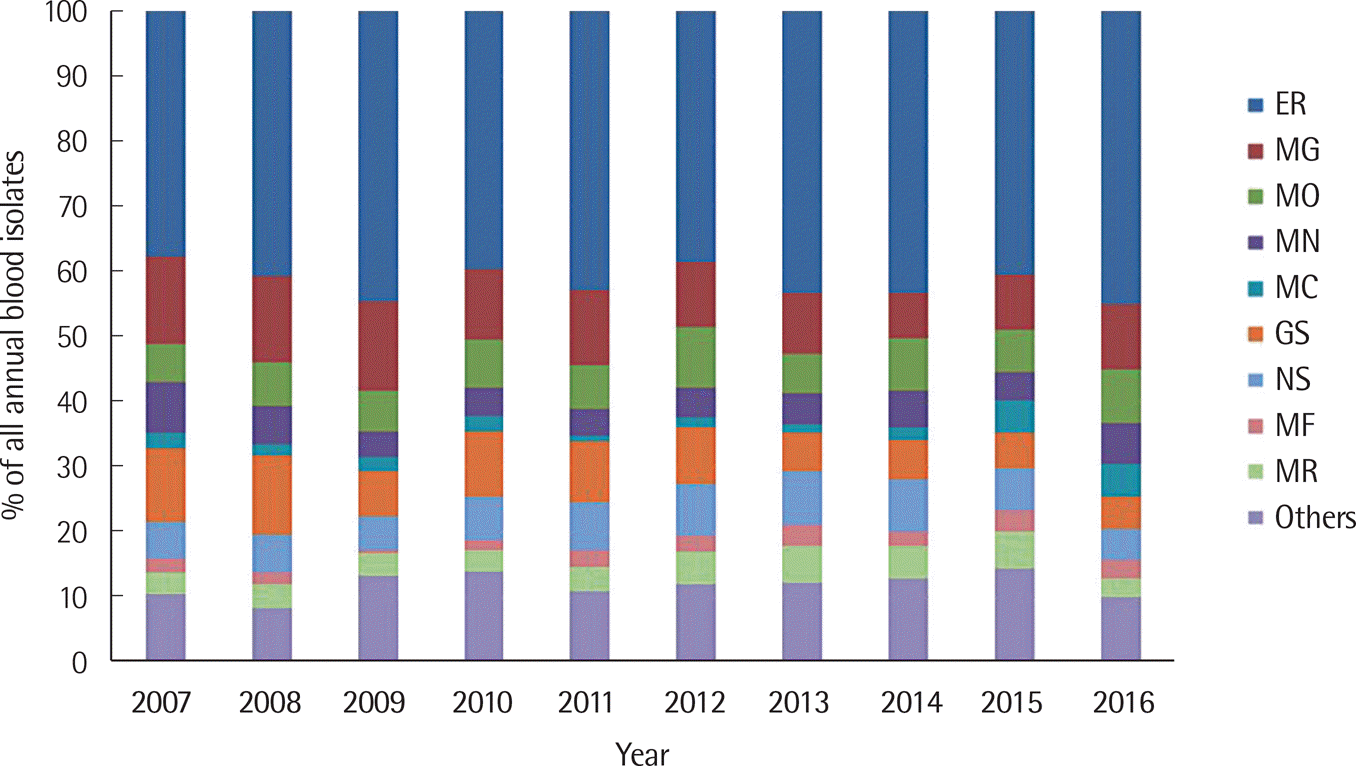 | Fig. 2.Proportion of isolated organisms from blood cultures according to department of medicine between 2007 and 2016. Abbreviations: ER, Emergency Room; MG, Medicine of Gastroenterology; MO, Medicine of Oncology; MN, Medicine of Nephrology; MC, Medicine of Cardiovascular; GS, General Surgery; NS, Neurological Surgery; MF, Medicine of Infectious Diseases; MR, Medicine of Respiratory. |
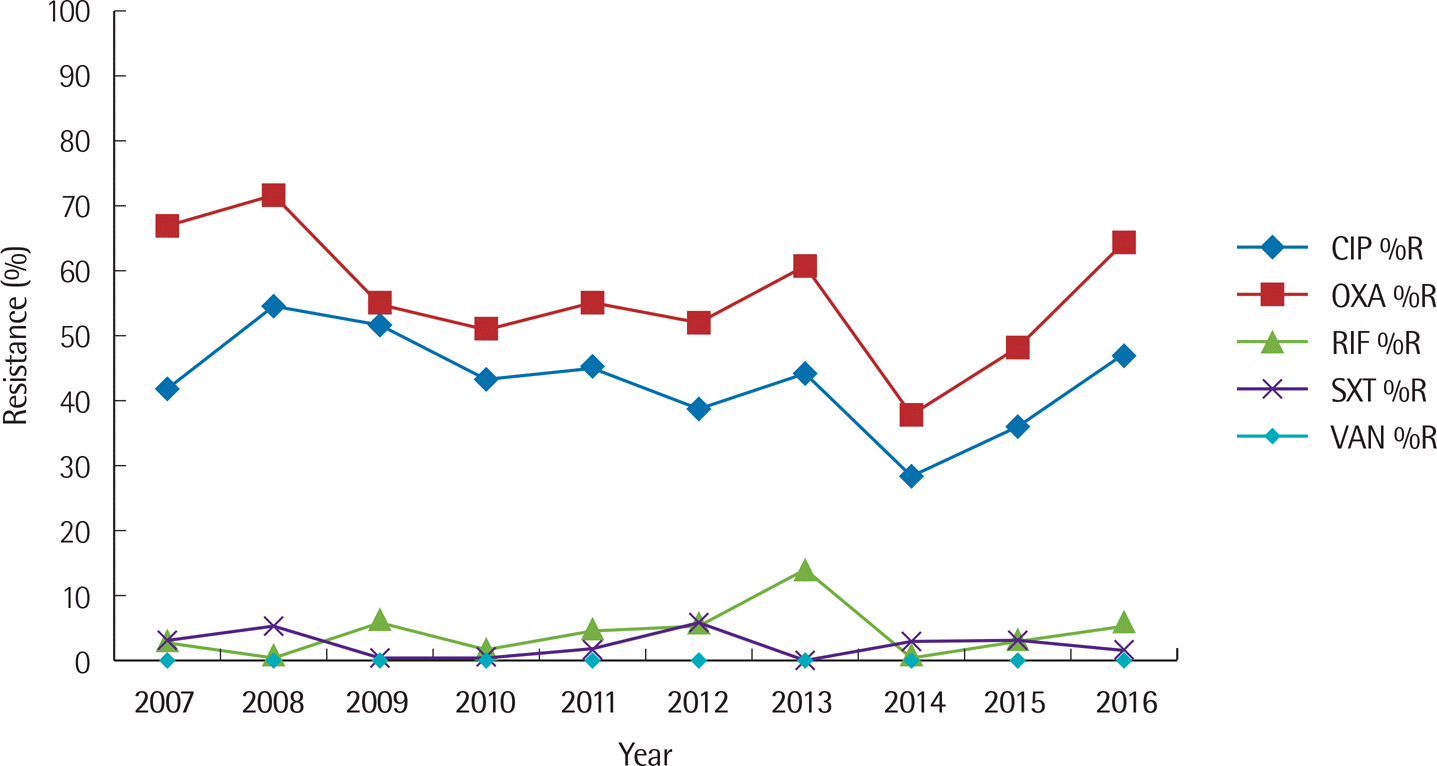 | Fig. 3.Ten-year trend of percent resistance to ciprofloxacin, oxacillin, rifampin, cotrimoxazole, and vancomycin for S. aureus. Abbreviations: CIP, ciprofloxacin; OXA, oxacillin; RIF, rifampin; SXT, cotrimoxazole; VAN, vancomycin; %R, % resistance. |
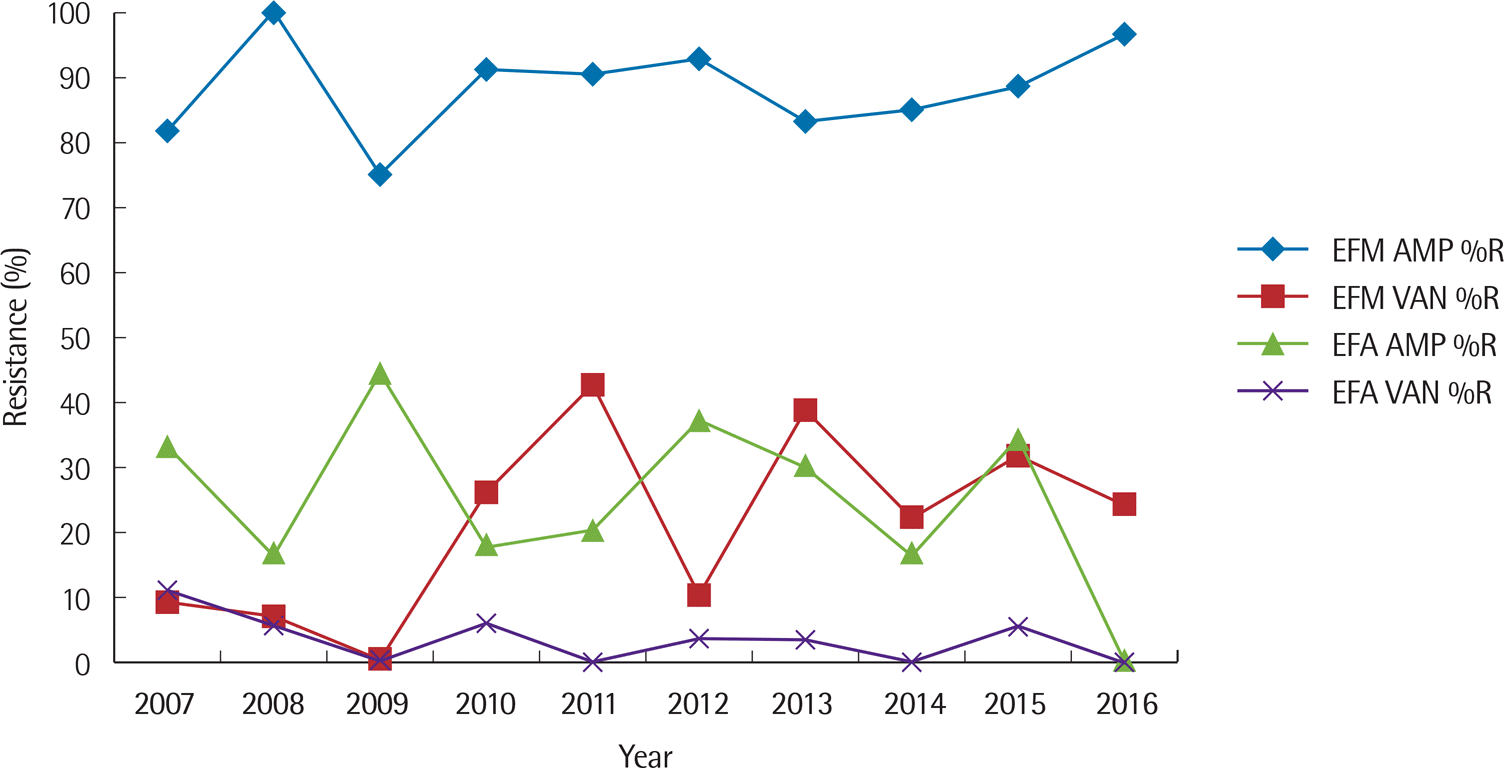 | Fig. 4.Ten-year trend of percent resistance to ampicillin and vancomycin for E. faecium and E. faecalis. Abbreviations: EFM, E. faecium; EFA, E. faecalis; AMP, ampicillin; VAN, vancomycin; %R, % resistance. |
 | Fig. 5.Ten-year trend of percent resistance to third-generation cephalosporins and fluoroquinolone for E. coli and K. pneumoniae. Abbreviations: ECO, E. coli; KPN, K. pneumoniae; 3rd cepha, third-generation cephalosporins; FQ, fluoroquinolone; %R, % resistance. |
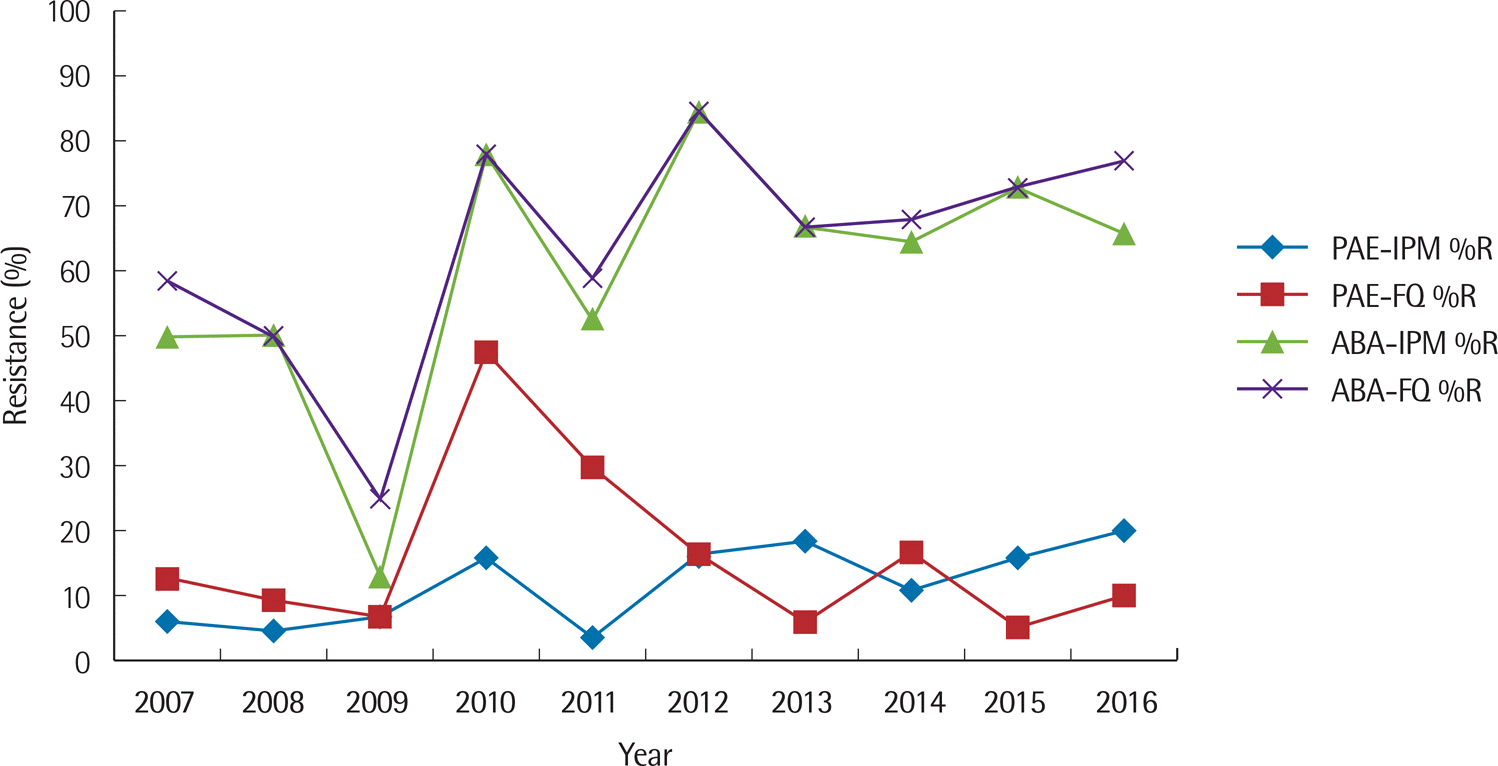 | Fig. 6.Ten-year trend of percent resistance to imipenem and fluoroquinolone for P. aeruginosa and A. baumannii. Abbreviations: PAE, P. aeruginosa; ABA, A. baumannii; IPM, imipenem; FQ, fluoroquinolone; %R, % resistance. |
Table 1.
Relatively common species of isolated bacteria by age group
Table 2.
Frequency of bacterial isolates from blood cultures




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download