Abstract
Purpose
Materials and Methods
Results
Figures and Tables
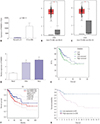 | Fig. 1FAM83H over-expression is associated with poor prognosis of CC. (A) FAM83H expression at mRNA level in CC tissues in TCGA database. (B) FAM83H expression at mRNA level in CC tissues in GEPIA database (match TCGA normal data). (C) FAM83H expression at mRNA level in CC tissues in GEPIA database (match TCGA normal and GTEx data). (D) Real-time Quantitative Reverse Transcription PCR used to measure the expression level of FAM83H in human CC cell lines (SiHa and HeLa). The expression of FAM83H was significantly higher in CC cells relative to control cells. (E) Kaplan-Meier curves of OS of CC patients in TCGA database. Patients were sub-grouped into high (n=152) or low (n=152) groups based on the median of FAM83H expression. p=0.027. (F) Kaplan-Meier curves of OS of CC patients in GEPIA database. Patients were sub-grouped into high (n=146) or low (n=146) groups based on the median of FAM83H expression. (G) Kaplan-Meier curves of OS of CC patients in the Human Protein Atlas database. Patients were sub-grouped into high (n=224) or low (n=67) groups based on best cut-off point (p=0.004). *p<0.05, **p<0.01. FAM83H, family with sequence similarity 83 member H; CC, cervical cancer; TCGA, The Cancer Genome Atlas; GEPIA, Gene Expression Profiling Interactive Analysis; OS, overall survival; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; TPM, transcripts per million; HR, hazard ratio. |
 | Fig. 2FAM83H expression in HeLa cells transfected with siRNA FAM83H 1# and siRNA FAM83H 2# were measured by Real-Time Quantitative Reverse Transcription PCR (A) and Western blotting (B and C). *p<0.01 compared to the si-con group. FAM83H, family with sequence similarity 83 member H; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. |
 | Fig. 3Knockdown of FAM83H inhibits growth and viability of HeLa cells. CCK-8 assay showing viability of transfected HeLa cells at 24, 48, 72, and 96 h. *p<0.05 compared to control, **p<0.001 compared to control. CCK-8, Cell-Counting Kit 8; FAM83H, family with sequence similarity 83 member H; OD, optical density. |
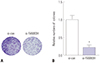 | Fig. 4Knockdown of FAM83H decreases colony formation of HeLa cells. (A) Representative images of colony formation assay (×2). The clones were dyed by 0.1% crystal violet. (B) Quantification of the colony numbers. *p<0.001 compared to control. FAM83H, family with sequence similarity 83 member H. |
 | Fig. 5Analysis of wound-healing (A; ×100) and transwell assay [B (×200) and C]. Cells were dyed by 0.1% crystal violet. In HeLa cells transfected with si-FAM83H, the invasion and migration were significantly decreased compared to the cells transfected with scrambled si-RNA (si-con). *p<0.001 compared to control. FAM83H, family with sequence similarity 83 member H. |
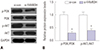 | Fig. 6The expression of PI3K/AKT pathway related proteins. After FAM83H silencing, the expression of p-AKT and p-PI3K were all reduced significantly. (A) The expression of PI3K pathway related proteins were determined by Western blot. (B) Quantification of the protein expression levels in (A). *p<0.001 compared to control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; FAM83H, family with sequence similarity 83 member H. |
Notes
AUTHOR CONTRIBUTIONS
Conceptualization: Chao Chen and Jia Zhou.
Data curation: Chao Chen and Hua-Feng Li.
Formal analysis: Yu-Jie Hu.
Investigation: Chao Chen, Hua-Feng Li, Yu-Jie Hu, Meng-Jie Jiang, and Qing-Sheng Liu.
Methodology: Chao Chen.
Project administration: Chao Chen and Jia Zhou.
Software: Hua-Feng Li and Yu-Jie Hu.
Supervision: Jia Zhou.
Validation: Meng-Jie Jiang and Qing-Sheng Liu.
Visualization: Chao Chen.
Writing—original draft: Chao Chen.
Writing—review & editing: Jia Zhou.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download