Abstract
The olfactory epithelium is capable of structural and functional recovery after injury through neurogenesis. Neurogenesis occurs via stem cells in the olfactory epithelium. Horizontal basal cells and globose basal cells in the basal layer of the epithelium have the characteristics of stem cells and progenitor cells of olfactory neurons. In order for the horizontal basal cells and globose basal cells to differentiate into olfactory neurons, distinct transcriptional factors are required at each stage. These transcription factors inhibit or synergize with each other or cells at each differentiation stage, regulating olfactory neurogenesis. Recently, the regulation of neurogenesis and development through epigenetic controls that change gene expression without changing the gene sequence have been studied. Studies of olfactory epithelium have helped to elucidate complex neurological systems including spinal cord and brain. In particular, features of neurogenesis will lead to medical advances in the treatment of central nervous diseases, which until this time have been considered impossible.
Figures and Tables
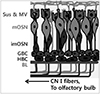 | Fig. 1Schematic illustration of the olfactory epithelium. Olfactory epithelium (OE) is a pseudostratified neuroepithelium, housing the cell bodies of mature olfactory sensory neurons (mOSN), as well as immature neurons (imOSN) produced from basal stem cells. Two populations of stem and progenitor cells, the globose basal cells (GBC) and the horizontal basal cells (HBC), support self-renewal of the OE, replacing neurons as needed. Glia-like sustentacular (Sus) and sensory microvillar (MV) cells are situated apically. Axons from OSNs exit the base of the OE and their fascicles project as the first cranial nerve (CN I) (Slightly modified from reference#1). |
 | Fig. 2Transcriptional regulation of cell specification in the olfactory epithelium. Progenitors including horizontal basal cells (HBCs) and globose basal cells (GBCs) require transcription factors (TFs) marked in green to be able to proliferate and survive. The neuronal lineage pathway begins with the expression of proneural TFs (such as Mash1 and Ngn1). Intermediate progenitors (IP) then differentiate under the control of differentiation TFs (such as Lhx2) to become immature olfactory sensory neuron (imOSN). imOSN subsequently undergo maturation by extension toward the apical (AL) and basal layer (BL) under the control of maturation TFs (such as OMP and O/E) to become mature sensory neurons (mOSN). Sox2/Pax6+progenitors can also choose a non-neuronal path to generate sustentacular (SUS) cells under the regulation of differentiation/maturation TFs (such as Steel and Hes1). Other non-neuronal derivatives of Sox2/Pax6+progenitors include Bowman's glands (BGs) and microvillar cells (MCs) specified by TFs marked in deep and light green respectively (Slightly modified from reference#44). |
Table 1
Studies of epigenetic factors in olfactory epithelium development

cKO: conditional knock-out, dKO: double knock-out, cOE: conditional overexpression, KD: knock-down, OE: olfactory epithelium, oNSCs: olfactory neural stem cells, IPs: intermediate progenitors, im: immature, m: mature, OSNs: olfactory sensory neurons, OR: olfactory receptor (Slightly modified from reference#44)
References
1. Choi R, Goldstein BJ. Olfactory epithelium: Cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope Investig Otolaryngol. 2018; 3:35–42.

2. Holbrook EH, Wu E, Curry WT, Lin DT, Schwob JE. Immunohistochemical characterization of human olfactory tissue. Laryngoscope. 2011; 121:1687–1701.

3. Farbman AI, Margolis FL. Olfactory marker protein during ontogeny: immunohistochemical localization. Dev Biol. 1980; 74:205–215.

4. Ronnett GV, Moon C. G proteins and olfactory signal transduction. Annu Rev Physiol. 2002; 64:189–222.

5. De Lorenzo AJ. Electron microscopic observations of the olfactory mucosa and olfactory nerve. J Biophys Biochem Cytol. 1957; 3:839–850.

8. Caggiano M, Kauer JS, Hunter DD. Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron. 1994; 13:339–352.

9. Calof AL, Chikaraishi DM. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 1989; 3:115–127.

10. Goldstein BJ, Schwob JE. Analysis of the globose basal cell compartment in rat olfactory epithelium using GBC-1, a new monoclonal antibody against globose basal cells. J Neurosci. 1996; 16:4005–4016.

11. Schwob JE, Huard JM, Luskin MB, Youngentob SL. Retroviral lineage studies of the rat olfactory epithelium. Chem Senses. 1994; 19:671–682.

12. Chen X, Fang H, Schwob JE. Multipotency of purified, transplanted globose basal cells in olfactory epithelium. J Comp Neurol. 2004; 469:457–474.

13. Mackay-Sim A, Kittel P. Cell dynamics in the adult mouse olfactory epithelium: a quantitative autoradiographic study. J Neurosci. 1991; 11:979–984.

14. Mahanthappa NK, Schwarting GA. Peptide growth factor control of olfactory neurogenesis and neuron survival in vitro: roles of EGF and TGF-beta s. Neuron. 1993; 10:293–305.

15. Satoh M, Takeuchi M. Induction of NCAM expression in mouse olfactory keratin-positive basal cells in vitro. Brain Res Dev Brain Res. 1995; 87:111–119.

16. Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007; 10:720–726.

17. Iwai N, Zhou Z, Roop DR, Behringer RR. Horizontal basal cells are multipotent progenitors in normal and injured adult olfactory epithelium. Stem Cells. 2008; 26:1298–1306.

18. Joiner AM, Green WW, McIntyre JC, Allen BL, Schwob JE, Martens JR. Primary Cilia on Horizontal Basal Cells Regulate Regeneration of the Olfactory Epithelium. J Neurosci. 2015; 35:13761–13772.

19. Guo Z, Packard A, Krolewski RC, Harris MT, Manglapus GL, Schwob JE. Expression of pax6 and sox2 in adult olfactory epithelium. J Comp Neurol. 2010; 518:4395–4418.

20. Schwob JE. Restoring olfaction: a view from the olfactory epithelium. Chem Senses. 2005; 30:Suppl 1. i131–i132.

21. Goldstein BJ, Fang H, Youngentob SL, Schwob JE. Transplantation of multipotent progenitors from the adult olfactory epithelium. Neuroreport. 1998; 9:1611–1617.

22. Suzuki J, Sakurai K, Yamazaki M, Abe M, Inada H, Sakimura K, et al. Horizontal Basal Cell-Specific Deletion of Pax6 Impedes Recovery of the Olfactory Neuroepithelium Following Severe Injury. Stem Cells Dev. 2015; 24:1923–1933.

23. Schwob JE, Youngentob SL, Mezza RC. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J Comp Neurol. 1995; 359:15–37.

24. Alan MS, James SJ, James ES. Neurogenesis in the Adult Olfactory Epithelium. In : Doty RL, editor. Handbook of Olfaction and Gustation. 3rd ed. John Wiley & Sons;2015.
25. Packard AI, Lin B, Schwob JE. Sox2 and Pax6 Play Counteracting Roles in Regulating Neurogenesis within the Murine Olfactory Epithelium. PLoS One. 2016; 11:e0155167.

26. Duggan CD, DeMaria S, Baudhuin A, Stafford D, Ngai J. Foxg1 is required for development of the vertebrate olfactory system. J Neurosci. 2008; 28:5229–5239.

27. Gordon MK, Mumm JS, Davis RA, Holcomb JD, Calof AL. Dynamics of MASH1 expression in vitro and in vivo suggest a non-stem cell site of MASH1 action in the olfactory receptor neuron lineage. Mol Cell Neurosci. 1995; 6:363–379.

28. Shaker T, Dennis D, Kurrasch DM, Schuurmans C. Neurog1 and Neurog2 coordinately regulate development of the olfactory system. Neural Dev. 2012; 7:28.

29. Packard A, Giel-Moloney M, Leiter A, Schwob JE. Progenitor cell capacity of NeuroD1-expressing globose basal cells in the mouse olfactory epithelium. J Comp Neurol. 2011; 519:3580–3596.

30. Berghard A, Hagglund AC, Bohm S, Carlsson L. Lhx2-dependent specification of olfactory sensory neurons is required for successful integration of olfactory, vomeronasal, and GnRH neurons. Faseb J. 2012; 26:3464–3472.

31. Yu Y, Ren W, Ren B. Expression of signal transducers and activator of transcription 3 (STAT3) determines differentiation of olfactory bulb cells. Mol Cell Biochem. 2009; 320:101–108.

32. Behrens M, Venkatraman G, Gronostajski RM, Reed RR, Margolis FL. NFI in the development of the olfactory neuroepithelium and the regulation of olfactory marker protein gene expression. Eur J Neurosci. 2000; 12:1372–1384.

33. Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000; 127:2323–2332.

34. Wittmann W, Iulianella A, Gunhaga L. Cux2 acts as a critical regulator for neurogenesis in the olfactory epithelium of vertebrates. Dev Biol. 2014; 388:35–47.

35. Donner AL, Episkopou V, Maas RL. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol. 2007; 303:784–799.

36. Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013; 12:15–30.

37. Collinson JM, Quinn JC, Hill RE, West JD. The roles of Pax6 in the cornea, retina, and olfactory epithelium of the developing mouse embryo. Dev Biol. 2003; 255:303–312.

38. Krolewski RC, Packard A, Jang W, Wildner H, Schwob JE. Ascl1 (Mash1) knockout perturbs differentiation of nonneuronal cells in olfactory epithelium. PLoS One. 2012; 7:e51737.

39. Murray RC, Navi D, Fesenko J, Lander AD, Calof AL. Widespread defects in the primary olfactory pathway caused by loss of Mash1 function. J Neurosci. 2003; 23:1769–1780.

40. Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002; 129:1871–1880.

41. Kudrycki K, Stein-Izsak C, Behn C, Grillo M, Akeson R, Margolis FL. Olf-1-binding site: characterization of an olfactory neuron-specific promoter motif. Mol Cell Biol. 1993; 13:3002–3014.

42. Lee AC, He J, Ma M. Olfactory marker protein is critical for functional maturation of olfactory sensory neurons and development of mother preference. J Neurosci. 2011; 31:2974–2982.

43. Baumeister H, Gronostajski RM, Lyons GE, Margolis FL. Identification of NFI-binding sites and cloning of NFI-cDNAs suggest a regulatory role for NFI transcription factors in olfactory neuron gene expression. Brain Res Mol Brain Res. 1999; 72:65–79.

44. Sokpor G, Abbas E, Rosenbusch J, Staiger JF, Tuoc T. Transcriptional and Epigenetic Control of Mammalian Olfactory Epithelium Development. Mol Neurobiol. 2018; 55:8306–8327.

45. Sokpor G, Xie Y, Rosenbusch J, Tuoc T. Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front Mol Neurosci. 2017; 10:243.

46. Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007; 55:201–215.

47. Bachmann C, Nguyen H, Rosenbusch J, Pham L, Rabe T, Patwa M, et al. mSWI/SNF (BAF) Complexes Are Indispensable for the Neurogenesis and Development of Embryonic Olfactory Epithelium. PLoS Genet. 2016; 12:e1006274.

48. Layman WS, McEwen DP, Beyer LA, Lalani SR, Fernbach SD, Oh E, et al. Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum Mol Genet. 2009; 18:1909–1923.

49. MacDonald JL, Gin CS, Roskams AJ. Stage-specific induction of DNA methyltransferases in olfactory receptor neuron development. Dev Biol. 2005; 288:461–473.

50. Andrews PJ, Poirrier AL, Lund VJ, Choi D. Safety of human olfactory mucosal biopsy for the purpose of olfactory ensheathing cell harvest and nerve repair: a prospective controlled study in patients undergoing endoscopic sinus surgery. Rhinology. 2016; 54:183–191.

51. Holbrook EH, Rebeiz L, Schwob JE. Office-based olfactory mucosa biopsies. Int Forum Allergy Rhinol. 2016; 6:646–653.

52. Choi D, Li D, Law S, Powell M, Raisman G. A prospective observational study of the yield of olfactory ensheathing cells cultured from biopsies of septal nasal mucosa. Neurosurgery. 2008; 62:1140–1144.

53. Feron F, Perry C, McGrath JJ, Mackay-Sim A. New techniques for biopsy and culture of human olfactory epithelial neurons. Arch Otolaryngol Head Neck Surg. 1998; 124:861–866.

54. Feron F, Perry C, Cochrane J, Licina P, Nowitzke A, Urquhart S, et al. Autologous olfactory ensheathing cell transplantation in human spinal cord injury. Brain. 2005; 128:2951–2960.

55. Mackay-Sim A, Feron F, Cochrane J, Bassingthwaighte L, Bayliss C, Davies W, et al. Autologous olfactory ensheathing cell transplantation in human paraplegia: a 3-year clinical trial. Brain. 2008; 131:2376–2386.

56. Lima C, Pratas-Vital J, Escada P, Hasse-Ferreira A, Capucho C, Peduzzi JD. Olfactory mucosa autografts in human spinal cord injury: a pilot clinical study. J Spinal Cord Med. 2006; 29:191–203.

57. Li Y, Carlstedt T, Berthold CH, Raisman G. Interaction of transplanted olfactory-ensheathing cells and host astrocytic processes provides a bridge for axons to regenerate across the dorsal root entry zone. Exp Neurol. 2004; 188:300–308.

58. Woodhall E, West AK, Chuah MI. Cultured olfactory ensheathing cells express nerve growth factor, brain-derived neurotrophic factor, glia cell line-derived neurotrophic factor and their receptors. Brain Res Mol Brain Res. 2001; 88:203–213.

59. Chung RS, Woodhouse A, Fung S, Dickson TC, West AK, Vickers JC, et al. Olfactory ensheathing cells promote neurite sprouting of injured axons in vitro by direct cellular contact and secretion of soluble factors. Cell Mol Life Sci. 2004; 61:1238–1245.

60. Holbrook EH, Leopold DA, Schwob JE. Abnormalities of axon growth in human olfactory mucosa. Laryngoscope. 2005; 115:2144–2154.

61. Duan D, Lu M. Olfactory mucosa: a rich source of cell therapy for central nervous system repair. Rev Neurosci. 2015; 26:281–293.

62. Lee JH, Goedert M, Hill WD, Lee VM, Trojanowski JQ. Tau proteins are abnormally expressed in olfactory epithelium of Alzheimer patients and developmentally regulated in human fetal spinal cord. Exp Neurol. 1993; 121:93–105.

63. Crino PB, Martin JA, Hill WD, Greenberg B, Lee VM, Trojanowski JQ. Beta-Amyloid peptide and amyloid precursor proteins in olfactory mucosa of patients with Alzheimer’s disease, Parkinson’s disease, and Down syndrome. Ann Otol Rhinol Laryngol. 1995; 104:655–661.

64. Mundinano IC, Caballero MC, Ordonez C, Hernandez M, DiCaudo C, Marcilla I, et al. Increased dopaminergic cells and protein aggregates in the olfactory bulb of patients with neurodegenerative disorders. Acta Neuropathol. 2011; 122:61–74.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download