1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015; 65:5–29.

2. Vaidya AP, Curtin JP. The follow-up of ovarian cancer. Semin Oncol. 2003; 30:401–412.

3. Cotte E, Glehen O, Mohamed F, Lamy F, Falandry C, Golfier F, et al. Cytoreductive surgery and intraperitoneal chemo-hyperthermia for chemo-resistant and recurrent advanced epithelial ovarian cancer: prospective study of 81 patients. World J Surg. 2007; 31:1813–1820.

4. Paik ES, Lee YY, Lee EJ, Choi CH, Kim TJ, Lee JW, et al. Survival analysis of revised 2013 FIGO staging classification of epithelial ovarian cancer and comparison with previous FIGO staging classification. Obstet Gynecol Sci. 2015; 58:124–134.

5. Mizuno M, Kajiyama H, Shibata K, Mizuno K, Kawai M, Nagasaka T, et al. Prognostic value of histological type in stage IV ovarian carcinoma: a retrospective analysis of 223 patients. Br J Cancer. 2015; 112:1376–1383.

6. Makar AP, Baekelandt M, Tropé CG, Kristensen GB. The prognostic significance of residual disease, FIGO substage, tumor histology, and grade in patients with FIGO stage III ovarian cancer. Gynecol Oncol. 1995; 56:175–180.

7. Mury D, Woelber L, Jung S, Eulenburg C, Choschzick M, Witzel I, et al. Prognostic and predictive relevance of CA-125 at primary surgery of ovarian cancer. J Cancer Res Clin Oncol. 2011; 137:1131–1137.

8. Rosendahl M, Høgdall CK, Mosgaard BJ. Restaging and survival analysis of 4036 ovarian cancer patients according to the 2013 FIGO classification for ovarian, fallopian tube, and primary peritoneal cancer. Int J Gynecol Cancer. 2016; 26:680–687.

9. Ayaru L, Ypsilantis PP, Nanapragasam A, Choi RC, Thillanathan A, Min-Ho L, et al. Prediction of outcome in acute lower gastrointestinal bleeding using gradient boosting. PLoS One. 2015; 10:e0132485.

10. Zhang Y, Zhou W, Yuan S, Yuan Q. Seizure detection method based on fractal dimension and gradient boosting. Epilepsy Behav. 2015; 43:30–38.

11. Prat A, Parera M, Peralta S, Perez-Benavente MA, Garcia A, Gil-Moreno A, et al. Nadir CA-125 concentration in the normal range as an independent prognostic factor for optimally treated advanced epithelial ovarian cancer. Ann Oncol. 2008; 19:327–331.

12. Riedinger JM, Wafflart J, Ricolleau G, Eche N, Larbre H, Basuyau JP, et al. CA 125 half-life and CA 125 nadir during induction chemotherapy are independent predictors of epithelial ovarian cancer outcome: results of a French multicentric study. Ann Oncol. 2006; 17:1234–1238.

13. Larose DT. Discovering knowledge in data: an introduction to data mining. Hoboken, NJ: John Wiley & Sons, Inc.;2005.
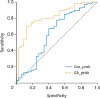
14. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988; 44:837–845.

15. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011; 12:2825–2830.
16. Zhu L, Luo W, Su M, Wei H, Wei J, Zhang X, et al. Comparison between artificial neural network and Cox regression model in predicting the survival rate of gastric cancer patients. Biomed Rep. 2013; 1:757–760.

17. Rutten MJ, Boldingh JH, Schuit E, Trum H, van Driel W, Mol BW, et al. Development and internal validation of a prognostic model for survival after debulking surgery for epithelial ovarian cancer. Gynecol Oncol. 2014; 135:13–18.

18. Barlin JN, Yu C, Hill EK, Zivanovic O, Kolev V, Levine DA, et al. Nomogram for predicting 5-year disease-specific mortality after primary surgery for epithelial ovarian cancer. Gynecol Oncol. 2012; 125:25–30.

19. Teramukai S, Ochiai K, Tada H, Fukushima M. Japan Multinational Trial Organization OC01-01. PIEPOC: a new prognostic index for advanced epithelial ovarian cancer--Japan Multinational Trial Organization OC01-01. J Clin Oncol. 2007; 25:3302–3306.

20. Chi DS, Palayekar MJ, Sonoda Y, Abu-Rustum NR, Awtrey CS, Huh J, et al. Nomogram for survival after primary surgery for bulky stage IIIC ovarian carcinoma. Gynecol Oncol. 2008; 108:191–194.

21. Clark TG, Stewart M, Rye T, Smyth JF, Gourley C. Validation of a new prognostic index for advanced epithelial ovarian cancer: results from its application to a UK-based cohort. J Clin Oncol. 2007; 25:5669–5670.

22. Tian C, Markman M, Zaino R, Ozols RF, McGuire WP, Muggia FM, et al. CA-125 change after chemotherapy in prediction of treatment outcome among advanced mucinous and clear cell epithelial ovarian cancers: a Gynecologic Oncology Group study. Cancer. 2009; 115:1395–1403.
23. Krivak TC, Tian C, Rose GS, Armstrong DK, Maxwell GL. A Gynecologic Oncology Group Study of serum CA-125 levels in patients with stage III optimally debulked ovarian cancer treated with intraperitoneal compared to intravenous chemotherapy: an analysis of patients enrolled in GOG 172. Gynecol Oncol. 2009; 115:81–85.

24. Rustin GJ. Can we now agree to use the same definition to measure response according to CA-125? J Clin Oncol. 2004; 22:4035–4036.

25. Lee CK, Friedlander M, Brown C, Gebski VJ, Georgoulopoulos A, Vergote I, et al. Early decline in cancer antigen 125 as a surrogate for progression-free survival in recurrent ovarian cancer. J Natl Cancer Inst. 2011; 103:1338–1342.

26. Mano A, Falcão A, Godinho I, Santos J, Leitão F, Oliveira C, et al. CA-125 AUC as a new prognostic factor for patients with ovarian cancer. Gynecol Oncol. 2005; 97:529–534.

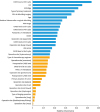
27. Zou J, Wang E. eTumorType, an algorithm of discriminating cancer types for circulating tumor cells or cell-free DNAs in blood. Genomics Proteomics Bioinformatics. 2017; 15:130–140.







 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download