INTRODUCTION
Enterococcus spp. are opportunistic pathogens that inhabit the intestines of many warm-blooded animals, including humans, and have been isolated from several sources such as food, plants, water, and soil [
1].
Enterococcus spp. can infect birds through various routes including the respiratory system, gastrointestinal tract, and integument, particularly in poultry environments where birds are hatched, reared, or processed. After infection, bacteria circulate in the bloodstream and form abscesses that cause arthritis, osteomyelitis, spondylitis, and femoral head necrosis in broiler and broiler breeder flocks [
2-
4]. Virulence of
Enterococcus spp. is associated with several genes including
fsr (regulator of
gelE expression),
efm (
E. faecium-specific cell wall adhesin),
esp (enterococcal surface protein),
cylA (hemolysin),
cad1 (pheromone cAD1 precursor lipoprotein),
ace (collagen-binding cell wall protein),
gelE (gelatinase),
asa1 (aggregation substance),
acm (surface-exposed antigen),
cpd1 (pheromone cPD1 lipoprotein),
efaAEfs (endocarditis-specific antigen), and
sagA (secreted antigen) [
5,
6]. These virulence factors are involved in attachment to target organs, colonization, invasion of host tissues, diminished host immune function, and the production of enzymes that increase the severity of infection and regulate extracellular production of toxins [
7-
9].
The use of antimicrobials to enhance the production of animals for human consumption of meat and eggs is leading to increased antimicrobial resistance worldwide.
Enterococcus spp. acquire antibiotic resistance via transfer of plasmids and transposons, chromosomal exchange, and spontaneous mutations [
6]. Consequently, the use of antibiotics for animal growth promotion exposes humans to antibiotic-resistant pathogens through food and contaminates the environment, which facilitates spreading.
Herein, we developed a multiplex polymerase chain reaction (PCR) method for simultaneous detection of 8 virulence genes in Enterococcus spp. and investigated potential virulence factors and antimicrobial resistance using E. faecium, E. faecalis, E. hirae, and E. gallinarum isolates from broiler chickens suffering lameness in 2 commercial poultry farms.
MATERIALS AND METHODS
Isolation of bacteria
Enterococcus spp. were isolated from various samples of 13 different flocks suffering from lameness in 2 commercial poultry farms over a period of 7 weeks in 2016. Broilers with lameness varied in age from 2 to 15 days. Swabs from liver, femur, and joint tissue were inoculated onto Enterococcosel agar and blood agar plates (BD Diagnostics, Germany) and cultivated at 37°C for 18–24 h. After the initial growth period, dark brown colonies on selective Enterococcosel agar medium were presumptively characterized as Enterococcus spp. and subcultured in brain heart infusion (BHI) agar for Enterococcus species-specific identification using 16s rRNA sequencing analysis and conventional biochemical testing, including Gram staining and Vitek2.
DNA extraction
Total genomic DNA was isolated from 80 different bacterial species using a QIAamp DNA mini kit (Qiagen, Germany) according to the manufacturer's instructions.
Oligonucleotide primer design
Specific primers were designed based on published
fsr (GenBank accession No. JN246675),
efm (GenBank accession No. AF097414),
esp (GenBank accession No. AH013271 and JF826520),
cylA (GenBank accession No. JQ794947 and KY613925),
cad1 (GenBank accession No. AF421355, NC017960, CP003504, and AOSM01000007),
ace (GenBank accession No. AF159247 and KY613927),
gelE (GenBank accession No D85393, KT598464, EU423275, and KY613931), and
asa1 (GenBank accession No. KT598461, KY613929, and U91527) gene sequences using OligoAnalyzer (Integrated DNA Technologies, USA). Primer specificity was confirmed by BLAST searches against the National Center for Biotechnology Information (NCBI) database (
https://www.ncbi.nlm.nih.gov/). The eight pairs of primers and amplicon sizes are listed in
Table 1.
Table 1
Sequences of oligonucleotide primers used to detect putative virulence genes in Enterococcus isolates

|
Virulence gene |
Gene description |
Primer |
Sequence (5′ to 3′) |
Amplicon size (bp) |
|
fsr
|
Regulator of gelE and sprE expression |
Forward primer |
CAA GGC ACT ATT TCT TAC TTA GG |
1,016 |
|
Reverse primer |
AGC GCA TAA ATC AAC CAA G |
|
efm
|
E. faecium-specific cell wall adhesin |
Forward primer |
GAA AAG TTG TCA GTC GTG G |
818 |
|
Reverse primer |
TGT TTG TGA CAA ACC TTC ATG |
|
esp
|
Enterococcal surface protein |
Forward primer |
CAT CTT TGA TTC TTG GTT GTC G |
695 |
|
Reverse primer |
GTT ATA GGT ACG TAT GTT GCA TCA |
|
cylA
|
Hemolysin |
Forward primer |
GAG TTA GAT GAA TAT GGT CAT GGT |
521 |
|
Reverse primer |
AGA AAC TAG CGA TGT AGG GTA ATA |
|
cad1
|
Pheromone cAD1 precursor lipoprotein |
Forward primer |
TTC CAA AAC TAC GCA CAA CA |
423 |
|
Reverse primer |
CTT TTT CAG CAG CAT TCA CTA ATT |
|
ace
|
Collagen-binding cell wall protein |
Forward primer |
ATA GAA ACG GAT TTC GGA ACA G |
298 |
|
Reverse primer |
TCA AAC TCG GCA AGT GAA ATA T |
|
gelE
|
Gelatinase |
Forward primer |
TAT GAC AAT GCT TTT TGG GAT G |
208 |
|
Reverse primer |
GCA CCC GAA ATA ATA TAA CCC |
|
asa1
|
Aggregation substance |
Forward primer |
AAC AAG CTT GGT CTG TGT ATC |
168 |
|
Reverse primer |
TCT TCC CCT TTC TTG TTA TGA AC |

Multiplex PCR
PCR assays (20 µL) were conducted using PCR premix (iNtRON Biotechnology, Seongnam, Korea). Each reaction mixture contained 2 µL bacterial genomic DNA, 0.1 µM of each primer for fsr, efm, esp, cylA, gelE, and asa1, 0.16 µM of each primer for cad1 and ace, 2.5 mM of each dNTP, 1.5 mM MgCl2, 10× reaction buffer, and 1 U Taq polymerase. The thermal cycler (Eppendorf, Hamburg, Germany) was programmed for initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and then final elongation at 72°C for 7 min. PCR products were analyzed under UV light after separation by 1.5% agarose gel electrophoresis.
Determining the multiplex PCR detection limit
The detection limit of multiplex PCR assays was determined by testing 5-fold serial dilutions of bacterial DNA of E. faecalis (field isolate) containing virulence genes fsr, esp, cylA, cad1, ace, gelE, and asa1. Similarly, the sensitivity for detecting the pheromone cAD1 precursor lipoprotein gene and E. faecium-specific cell wall adhesin gene was tested using E. hirae (field isolate) possessing the cAD1 gene and E. faecium (field isolate) harboring efm and cAD1 genes, respectively.
Sequencing method
To confirm the exact virulence genes, all PCR products amplified by multiplex and simplex PCR were purified using a QIAquick Gel Extraction Kit (Qiagen) and sequencing was performed by Cosmogenetech Co. (Korea). Nucleotide sequences were analyzed by BLAST searching against the NCBI database.
Screening of virulence genes in Enterococcus spp.
Multiplex PCR was used to analyze eight virulence genes in 26 E. faecalis, 25 E. faecium, 20 E. hirae, and 9 E. gallinarum isolates from liver, femur and joint tissues of broiler chickens suffering lameness.
Antibiotic susceptibility testing
The 16 antimicrobial minimal inhibitory concentrations (MICs) of Enterococcus isolates were determined using the Sensititre automated system (Trek Diagnostic Systems, USA) according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2013). Ampicillin, chloramphenicol, ciprofloxacin, daptomycin, erythromycin, florfenicol, gentamicin, kanamycin, linezolid, quinupristin-dalfopristin, salinomycin, streptomycin, tetracycline, tigecycline, tylosin, and vancomycin antibiotics were tested with Sensititre plates. E. faecalis ATCC 29212 and S. aureus ATCC 25923 strains were used as quality controls.
DISCUSSION
Many
Enterococcus spp. from various sources including humans, animals, birds, and the environment are generally considered harmless, although some can become opportunistic pathogens by acquiring antibiotic resistance and putative virulence genes from other bacteria through horizontal gene transfer [
10-
15]. Herein, the pathogenicity of isolates was evaluated by screening for the presence of virulence factors, and a multiplex PCR method was developed for the simultaneous detection of 8 enterococcal virulence genes (
fsr,
efm,
esp,
cylA,
cad1,
ace,
gelE, and
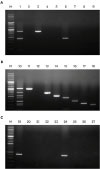
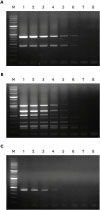
asa1). All primer pairs were validated using simplex PCR before being employed in multiplex PCR and only 1 amplified product of the expected size was observed in each simplex PCR as expected. In addition, we confirmed that each primer pair amplified the correct putative virulence gene by DNA sequencing.
To evaluate the sensitivity of multiplex PCR, DNA isolated from E. faecium harboring efm and cAD1 genes, E. faecalis harboring fsr, esp, cylA, cad1, ace, gelE, and asa1 genes, and E. hirae harboring the cad1 gene was tested at different concentrations. The detection limits of E. faecium, E. faecalis, and E. hirae were 64.0 pg/µL, 320.0 pg/µL, and 1.6 ng/µL DNA, respectively. Moreover, multiplex PCR was carried out using 5 µL heat-treated suspensions from individual colonies as DNA template, and the sensitivity results mirrored the detection limits obtained using genomic DNA isolated with a DNA extraction kit (data not shown). Thus, the multiplex PCR method for the 8 virulence genes was shown to be a reliable and rapid alternative for phenotypic testing and simplex PCR.
Furthermore, the multiplex PCR method was applied to screen 26
E. faecium, 25
E. faecalis, 20
E. hirae, and 9
E. gallinarum isolates from broiler chickens suffering lameness for the presence of these virulence genes, and the results showed that the assay could detect all 8 virulence genes in
Enterococcus spp. isolates. Additionally, all 26
E. faecium isolates were positive for
efm, whereas all
E. faecalis isolates were positive for ace. Among the virulence genes analyzed, the sex pheromone cAD1 precursor lipoprotein gene
cad1, which is related to the secretion of signaling molecules for intercellular communication, was detected in all 26
E. faecium, 25
E. faecalis, and 20
E. hirae isolates. Interestingly, aggregation substances encoded by sex pheromone plasmids are known to mediate aggregation between bacterial cells and facilitate transfer of plasmids [
16]. Hemolysin secreted by bacteria damages cell membranes and facilitates the infection process [
16,
17].
The distribution of
asa1 and
cylA among
E. faecalis isolates was 44% and 16%, respectively. The
gelE gene is believed to enhance the survivability of
Enterococcus spp. in extra-intestinal environments, and this gene was found in 88% of the 25
E. faecalis isolates. Gelatinase activity is known to be co-controlled by
gelE and
fsr genes, and lack of
fsr affects the production of gelatinase [
18,
19]. The
fsr gene product, which hydrolyzes gelatin, casein, hemoglobin, and other bioactive peptides, was detected in 64% of
E. faecalis isolates. Among
E. faecalis isolates harboring the
esp gene, which contributes to enterococcal biofilm formation, resistance to environmental stresses, and adhesion to eukaryotic cells, only 1 of 25 isolates was positive fpr
esp. Meanwhile,
fsr,
esp,
cylA,
ace,
gelE, and
asa1 genes were not detected in 26
E. faecium isolates, and
fsr,
esp,
cylA,
cad1,
ace,
gelE, and
asa1 genes were detected in 2 of 25
E. faecalis isolates. No virulence factors were found in any of the 9
E. gallinarum isolates.
Enterococcus spp. are widespread, resistant to various antimicrobial agents, and have the ability to rapidly acquire resistance to various antimicrobial agents [
5,
20]. Herein, all 49
Enterococcus isolates were resistant to tigecycline, followed by ciprofloxacin (73.5%), quinupristin/dalfopristin (55.1%), tetracycline (49.0%), daptomycin (42.9%), erythromycin (36.7%), and tylosin (32.7%). Resistance to linezolid and vancomycin was not observed in any of the
E. faecium,
E. faecalis,
E. hirae, or
E. gallinarum isolates. Among the 49
Enterococcus isolates, 22
E. faecium isolates were resistant to ciprofloxacin (100.0%) and daptomycin (77.3%).
Moreover, E. faecalis displayed 95.5%, 63.8%, 54.6%, and 50.0% resistance to high levels of quinupristin/dalfopristin, tetracycline, ciprofloxacin, and erythromycin, respectively. Additionally, 66.7% of E. hirae isolates were resistant to tylosin, ciprofloxacin, and erythromycin. Resistance to daptomycin, tetracycline, and tigecycline was 100% in all E. gallinarum isolates.
Comparison of antimicrobial agents revealed that the frequency of resistance to daptomycin and ciprofloxacin was much higher in E. faecium (77.3% and 100.0%, respectively) than in E. faecalis (4.6% and 54.6%, respectively). Similarly, resistance to erythromycin, streptomycin, quinupristin/dalfopristin, and tetracycline was higher in E. faecalis (50.0%, 36.4%, 95.5%, and 63.8%, respectively) than E. faecium (22.7%, 9.1%, 22.7%, and 31.8%, respectively). Multidrug resistance was observed in the majority of isolates, and the prevalence of multidrug resistance (resistant to at least 3 antimicrobials) was 19/22 (86.4%) for E. faecium, 20/22 (90.9%) for E. faecalis, 2/3 (66.7%) for E. hirae, and 2/2 (100.0%) for E. gallinarum.
To the best of our knowledge, this report is the first to provide detailed antibiotic resistance patterns for Enterococcus spp. isolated from commercial broiler chickens suffering lameness. The developed multiplex PCR method may have diagnostic value for the reliable, cost- and time-effective detection of 8 putative virulence genes in Enterococcus spp. simultaneously using a single assay.








 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download