INTRODUCTION
Epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (EGFR-TKIs) have profoundly improved available management strategies for
EGFR-mutated non-small cell lung cancer (NSCLC) by achieving a superior response rate, progression-free survival (PFS) rate, and quality of life than various available cytotoxic agents.
1 Therefore, they have become the established standard treatment for patients with
EGFR-mutant NSCLC;
23 nevertheless, these patients typically exhibit acquired resistance to EGFR-TKIs (including gefitinib, erlotinib, and afatinib) after a median period of 1 year. Of these, approximately 60% develop a T790M gatekeeper mutation in the
EGFR exon 20 kinase domain.
456
Osimertinib was the first oral, irreversible EGFR-TKI to receive approval from the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) to be used as a treatment for advanced
EGFR-mutant NSCLC cases exhibiting acquired
EGFR T790M resistance mutations.
7 A previous (AURA3) study demonstrated that, compared to treatment with chemotherapy, osimertinib treatment significantly improved PFS time (median PFS, 4.4 vs. 10.1 months;
p<0.001) and objective response rate (ORR) (31% vs. 71%,
p<0.001) of patients with T790M-positive advanced NSCLC (after previous first-line EGFR-TKI treatment).
8
Therefore, the introduction of osimertinib has rendered the ability of clinicians to detect T790M mutations essential in the clinical setting, to ensure that patients receive the most appropriate treatment available.
9 However, it is technically difficult to perform and collect an adequate sample volume from lung tissue biopsies; therefore, rebiopsy can be a clinical barrier to assessing a patient's
EGFR mutation status. Moreover, obtaining a new tissue sample at the time of progression is often invasive, expensive, more technically difficult than an initial tissue biopsy, and sometimes infeasible.
10
Recently, noninvasive plasma
EGFR genotyping emerged as a novel method to detect
EGFR mutations, including T790M. In fact, the US FDA has now approved the use of osimertinib in NSCLC cases confirmed to harbor an
EGFR T790M mutation via noninvasive plasma
EGFR genotyping, whether or not tumor tissue is available for screening using standard methods.
11 Some genotyping platforms for analyzing circulating tumor DNA (ctDNA) have already received regulatory approval and been introduced into the clinical setting (for use in combination with retrospective and prospective validation methods), and many more are likely to do so in the near future. Current methods to detect
EGFR mutations via various platforms include direct and next-generation sequencing, the “scorpion” amplified refractory mutation system, and polymerase chain reaction (PCR) techniques including peptide nucleic acid clamping, droplet digital PCR, and beads, emulsions, amplification and magnetics (BEAMing) PCR. Each of these various platforms has a range of advantages and disadvantages, and detects
EGFR mutations with varying sensitivity and specificity in patients who are treatment-naïve, compared to those who exhibit acquired resistance.
12 In particular, liquid biopsies to detect T790M mutations are essential in cases of progressed
EGFR-mutated NSCLC (after treatment with first or second-generation EGFR-TKIs), for which tumor tissue is unavailable or scanty.
13
The recently conducted ASTRIS (
NCT02474355) trial was an open-label, single-arm, multinational, real-world treatment study that investigated the safety and efficacy of using osimertinib to treat patients with T790M-positive advanced NSCLC, who had previously been administered a first- and/or second-generation EGFR-TKI.
14 In the present study, we aimed to investigate the success rate of tissue rebiopsies that were conducted, and the incidence of T790M mutations in tissue and plasma samples that were collected upon NSCLC progression from patients enrolled in the ASTRIS study. Furthermore, we evaluated whether the efficacy of osimertinib treatment was associated with the detection of T790M mutations in patient tissue and/or plasma samples.
Go to :

RESULTS
Patient population
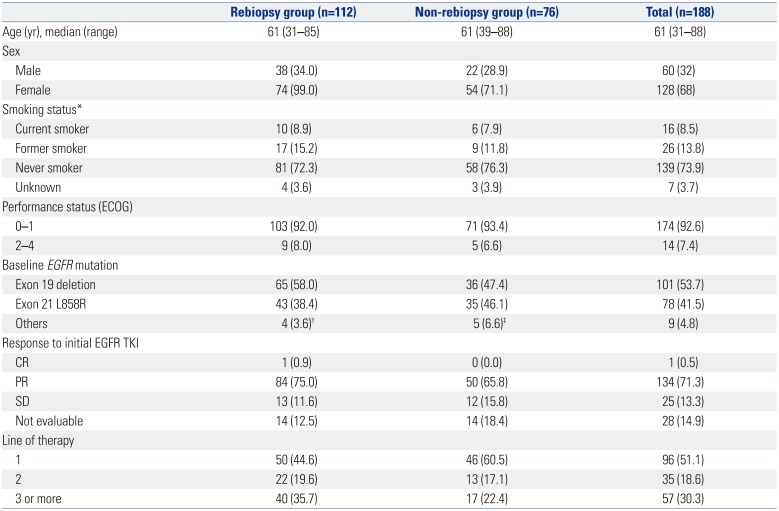
A total of 188 patients with NSCLC were screened for the ASTRIS study at Yonsei Cancer Center between April 2016 to December 2016 (
Table 1). Clinical data was based on a data cutoff from June 1, 2017. The median age of patients at study entry was 61 years (range 31–88 years), and all patients were histologically confirmed to exhibit adenocarcinoma. At the time of initial diagnosis, 53.7% (n=101) and 41.5% (n=78) of patients were found to harbor activating
EGFR exon 19 deletion and exon 21 L858R mutations, respectively, while 4.8% (n=9) patients instead exhibited rare
EGFR mutations. Most ASTRIS patients responded to the initial administration of EGFR-TKIs [ORR, 71.8%; 95% confidence interval (CI), 65.0–77.8%]. Only 51.1% of patients were exposed to one line of EGFR-TKI. A total of 23.4% (n=44) of patients were treated with two or more EGFR-TKIs, including seven patients who were administered another third-generation EGFR-TKI, olmutinib.
Table 1
Demographic and Clinical Characteristics of the Study Patients
|
Rebiopsy group (n=112) |
Non-rebiopsy group (n=76) |
Total (n=188) |
|
Age (yr), median (range) |
61 (31–85) |
61 (39–88) |
61 (31–88) |
|
Sex |
|
|
|
|
Male |
38 (34.0) |
22 (28.9) |
60 (32) |
|
Female |
74 (99.0) |
54 (71.1) |
128 (68) |
|
Smoking status*
|
|
|
|
|
Current smoker |
10 (8.9) |
6 (7.9) |
16 (8.5) |
|
Former smoker |
17 (15.2) |
9 (11.8) |
26 (13.8) |
|
Never smoker |
81 (72.3) |
58 (76.3) |
139 (73.9) |
|
Unknown |
4 (3.6) |
3 (3.9) |
7 (3.7) |
|
Performance status (ECOG) |
|
|
|
|
0–1 |
103 (92.0) |
71 (93.4) |
174 (92.6) |
|
2–4 |
9 (8.0) |
5 (6.6) |
14 (7.4) |
|
Baseline EGFR mutation |
|
|
|
|
Exon 19 deletion |
65 (58.0) |
36 (47.4) |
101 (53.7) |
|
Exon 21 L858R |
43 (38.4) |
35 (46.1) |
78 (41.5) |
|
Others |
4 (3.6)†
|
5 (6.6)‡
|
9 (4.8) |
|
Response to initial EGFR TKI |
|
|
|
|
CR |
1 (0.9) |
0 (0.0) |
1 (0.5) |
|
PR |
84 (75.0) |
50 (65.8) |
134 (71.3) |
|
SD |
13 (11.6) |
12 (15.8) |
25 (13.3) |
|
Not evaluable |
14 (12.5) |
14 (18.4) |
28 (14.9) |
|
Line of therapy |
|
|
|
|
1 |
50 (44.6) |
46 (60.5) |
96 (51.1) |
|
2 |
22 (19.6) |
13 (17.1) |
35 (18.6) |
|
3 or more |
40 (35.7) |
17 (22.4) |
57 (30.3) |

Rebiopsy feasibility after EGFR-TKI resistance acquisition
Among the ASTRIS cohort of 188 patients, 112 (59.5%) underwent rebiopsy. Rebiopsy was performed after the progression upon first- or second-generation EGFR-TKIs. Conversely, 76 (40.4%) patients did not undergo rebiopsy for various reasons; for example, in 37 cases, the treating physician decided against rebiopsy (
Fig. 1). Among them, 20 patients did not undergo a rebiopsy (as per the ASTRIS study guidelines), as their plasma samples had already been detected to harbor T790M mutations. In addition, 17 patients whose plasma samples were initially confirmed to be T790M-negative were managed by a “treatment-beyond-progression” strategy using previously administered EGFR-TKIs based on the treating physician's decision, and therefore, did not undergo a tissue rebiopsy. Moreover, 28 patients exhibited tumors that were inaccessible for rebiopsy, most (n=23) of which comprised small lung metastatic nodules, and four and one patient displayed only intracranial and skeletal cancer progression, respectively. Finally, five patients refused to undergo rebiopsy, and a further five were not able to undergo the rebiopsy procedure due to their poor performance status.
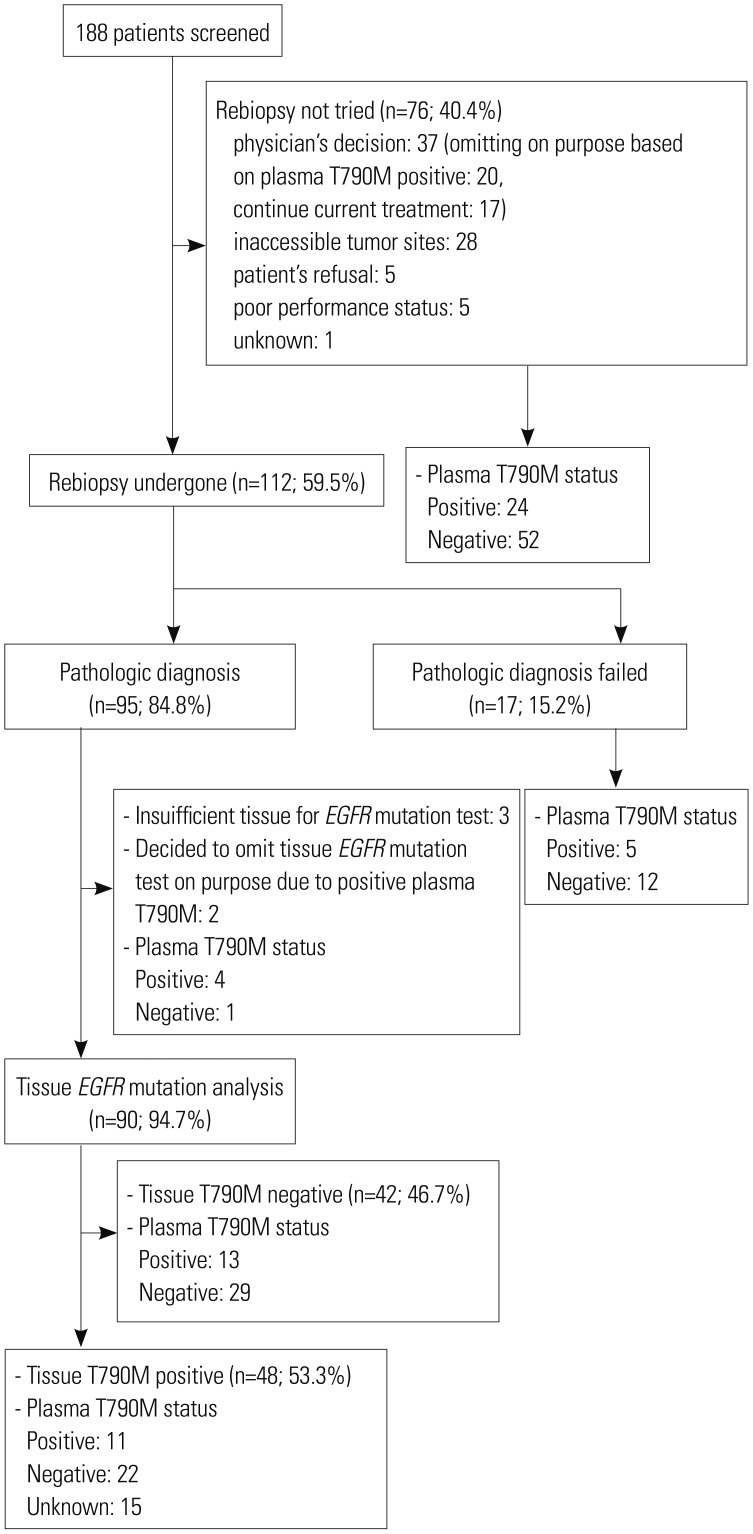 | Fig. 1CONSORT diagram. EGFR, epidermal growth factor receptor.
|
Of the 112 patients who did undergo rebiopsy, 18 had to undergo multiple biopsies to collect sufficient tumor tissue to enable EGFR mutation testing. In fact, two patients required a third biopsy, and furthermore, four patients failed to receive a valid mutation test result even after multiple biopsies. Overall, the rate of successful pathological confirmation of the rebiopsied tissues was 84.8% (95/112); however, in three of 95 cases, inadequate tissue was collected from the rebiopsy to complete EGFR mutation test. Therefore, excluding these and two other patients who underwent successful rebiopsy, but did not undergo EGFR mutation testing after T790M mutations, were detected in their plasma samples, and the overall success rate of tissue rebiopsy and subsequent EGFR mutation analysis among the ASTRIS cohort was 81.8% (90/110).
Among 88 patients who had undergone rebiopsy and could provide the information on initial biopsy, initial biopsy site were lung (n=82), pleural effusion (n=4), and neck lymph node (n=2). Among 82 patients whose initial biopsy site were lung, 67.1% of patients received rebiopsy in lung (n=55). For patients who had been diagnosed with pleural effusion sample only, rebiopsy with thoracentesis was done in one case and other rebiopsy targets included lung (n=2) and neck lymph node (n=1). For two patients who had undergone neck lymph node biopsy for initial diagnosis, one patient got neck lymph node biopsy and the other one underwent thoracentesis.
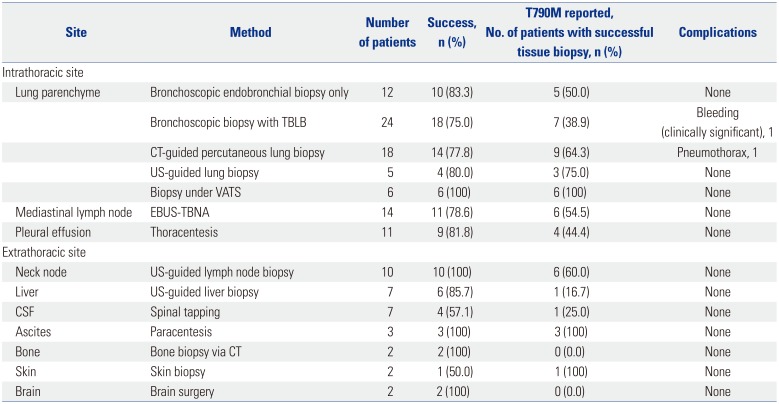
Rebiopsy methods, and success and complication rates
Various methods were used for rebiopsy patients (
Table 2). Of the 112 patients who underwent rebiopsy, 49 (43.8%) were subjected to bronchoscopy, including endobronchial biopsy, fluoroscopy-guided transbronchial lung biopsy (TBLB), or endobronchial ultrasound (EBUS)-transbronchial needle aspiration (TBNA). Of these methods, bronchoscopy with fluoroscopy- guided TBLB was most commonly used (n=24). The tumor acquisition rate achieved via this method and endobronchial biopsy method was 75.0% and 83.3%, respectively, and all methods were successfully employed to produce valid
EGFR mutation-test results. However, in one patient, all three bronchoscopic procedures were employed, and yet an appropriate tumor tissue sample could not be collected.
Table 2
Rebiopsy Methods and Their Success Rate and Complications (n=112)
|
Site |
Method |
Number of patients |
Success, n (%) |
T790M reported, No. of patients with successful tissue biopsy, n (%) |
Complications |
|
Intrathoracic site |
|
|
|
|
|
|
Lung parenchyme |
Bronchoscopic endobronchial biopsy only |
12 |
10 (83.3) |
5 (50.0) |
None |
|
Bronchoscopic biopsy with TBLB |
24 |
18 (75.0) |
7 (38.9) |
Bleeding (clinically significant), 1 |
|
CT-guided percutaneous lung biopsy |
18 |
14 (77.8) |
9 (64.3) |
Pneumothorax, 1 |
|
US-guided lung biopsy |
5 |
4 (80.0) |
3 (75.0) |
None |
|
Biopsy under VATS |
6 |
6 (100) |
6 (100) |
None |
|
Mediastinal lymph node |
EBUS-TBNA |
14 |
11 (78.6) |
6 (54.5) |
None |
|
Pleural effusion |
Thoracentesis |
11 |
9 (81.8) |
4 (44.4) |
None |
|
Extrathoracic site |
|
|
|
|
|
|
Neck node |
US-guided lymph node biopsy |
10 |
10 (100) |
6 (60.0) |
None |
|
Liver |
US-guided liver biopsy |
7 |
6 (85.7) |
1 (16.7) |
None |
|
CSF |
Spinal tapping |
7 |
4 (57.1) |
1 (25.0) |
None |
|
Ascites |
Paracentesis |
3 |
3 (100) |
3 (100) |
None |
|
Bone |
Bone biopsy via CT |
2 |
2 (100) |
0 (0.0) |
None |
|
Skin |
Skin biopsy |
2 |
1 (50.0) |
1 (100) |
None |
|
Brain |
Brain surgery |
2 |
2 (100) |
0 (0.0) |
None |

Twenty-three patients underwent percutaneous lung biopsy. The majority (78.3%, 18/23) of these patients were subjected to CT-guided percutaneous needle biopsy, with a success rate of 77.8% (14/18). Tumor acquisition rate of 100% was observed in neck lymph node biopsy (n=10), video-assisted thoracoscopic surgery (n=6), and brain surgery (n=2). In addition, a high pathological confirmation rate was achieved using various effusion sample types (i.e., pleural effusion, 81.8%, 9/11; ascites, 100%, 3/3).
Only one case each of significant bleeding (2.0%, 1/49) and pneumothorax (5.5%, 1/18) was observed among the rebiopsied patients, after bronchoscopy and CT-guided percutaneous lung biopsy, respectively. No clinically significant complications resulted from procedures that targeted non-lung tissues.
EGFR mutation test results
Of the 90 patients who underwent rebiopsy and received valid EGFR mutation test results, 53.3% (48/90) were found to harbor T790M mutation. Notably, the detection rate of tissue T790M mutation in all patients who progressed first or second-generation EGFR-TKIs was only 25.5% (48/188). Endobronchial biopsy, fluoroscopy-guided TBLB, and CT-guided percutaneous lung biopsy methods were used to identify EGFR T790M mutations in 50.0, 38.9, and 64.3% of applied cases, respectively. Similarly, EGFR T790M mutations were identified in all six patients who underwent rebiopsy via VATS, and all three patients whose malignant ascites was sampled via paracentesis. In contrast, T790M mutation was only detected in 16.7% and 25.0% of liver tissue and cerebrospinal fluid samples, respectively, and was not identified in any rebiopsied brain-tumor nor bone tissue samples.
Agreement between EGFR genotyping results generated using tissue and plasma samples
Plasma samples were collected in all 76 patients who were not rebiopsied, and genetic testing detected T790M mutations in 24 (31.6%) cases. Of the 188 patients in the ASTRIS cohort, 75 provided matched tumor tissue and plasma samples, whereas 98 and 15 patients only provided plasma or tissue samples, respectively, for
EGFR genotyping. A comparison of these matched samples revealed the positive predictive value of plasma T790M mutation screening to be 45.8% (11/24), and the false-positive rate among screened plasma samples to be 54.2% (13/24) (
Table 3).
Table 3
Comparison of T790M Status between Plasma and Tissue Samples (n=188)

|
Plasma T790M+ (n=57) |
Plasma T790M- (n=116) |
Plasma test not performed (n=15) |
|
Tissue T790M+ (n=48) |
11 |
22 |
15 |
|
Tissue T790M- (n=42) |
13 |
29 |
0 |
|
Tissue EGFR test not performed (n=98) |
33 |
65 |
0 |

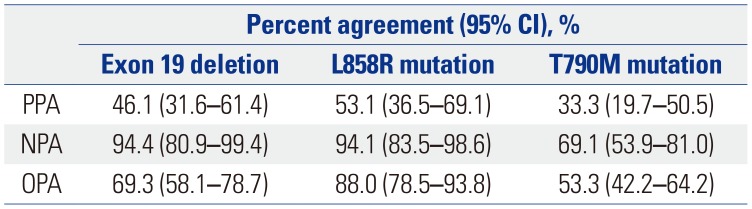
The overall PPA (sensitivity), NPA (specificity), and OPA of the plasma with tissue T790M-screening results was 33.3% (95% CI, 19.7–50.5%), 69.1% (95% CI, 53.9–81.0%), and 53.3% (95% CI, 42.2–64.2%), respectively. Notably, the utilized PNA-clamping genotyping method detected the activating
EGFR mutations (i.e., the
EGFR 19 deletion and L858R mutation) more effectively. For example, OPA and NPA between plasma and tissue screening for L858R mutation was 88.0% (95% CI, 78.5–93.8%) and 94.1% (95% CI, 83.5–98.6%), respectively (
Table 4).
Table 4
Percent Agreement of Plasma Assays (PANAMutyper™) with Tissue Genotype (PNAClamp™) as Reference Standard for EGFR Exon 19 Deletion, L858R Mutation, and T790M Mutation at the Time of Progression Upon EGFR-TKIs (n=75)
|
Percent agreement (95% CI), % |
|
Exon 19 deletion |
L858R mutation |
T790M mutation |
|
PPA |
46.1 (31.6–61.4) |
53.1 (36.5–69.1) |
33.3 (19.7–50.5) |
|
NPA |
94.4 (80.9–99.4) |
94.1 (83.5–98.6) |
69.1 (53.9–81.0) |
|
OPA |
69.3 (58.1–78.7) |
88.0 (78.5–93.8) |
53.3 (42.2–64.2) |

Association of osimertinib efficacy with tissue and/or plasma EGFR T790M mutation statuses
ORR among the 88 patients who were eligible for, and were administered treatment with osimertinib as part of the ASTRIS trial, was 44.3% (95% CI, 34.4–57.2%) (
Table 5). Notably, ORR was highest (57.8%) in patients who exhibited T790M-positive tumor tissue samples, modest (35.2%) in those who exhibited T790M-positive plasma samples, and lowest (7.6%) in those who exhibited a plasma samples that was positive, but a tissue samples that was negative for T790M mutation. Importantly, ORR of patients who exhibited T790M-positive tumor-tissue samples was greater than 50%, (57.8% in those with T790M-positive tissue, 54.5% in T790M-positive tissue/plasma, and 52.6% in T790M-positive tissue/T790M-negative plasma samples), regardless of their plasma test results. Likewise, the overall DCR was 78.4% (95% CI, 68.6–85.8%) among the 88 patients treated with osimertinib, but 100% among patients who provided T790M-positive tissue/T790M-negative plasma samples.
Table 5
Response Summary of Osimertinib according to the Source of T790M Status
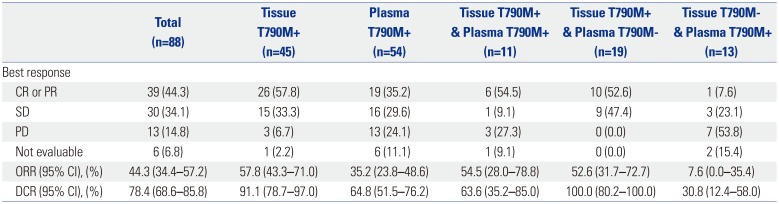
|
Total (n=88) |
Tissue T790M+ (n=45) |
Plasma T790M+ (n=54) |
Tissue T790M+ & Plasma T790M+ (n=11) |
Tissue T790M+ & Plasma T790M- (n=19) |
Tissue T790M− & Plasma T790M+ (n=13) |
|
Best response |
|
|
|
|
|
|
|
CR or PR |
39 (44.3) |
26 (57.8) |
19 (35.2) |
6 (54.5) |
10 (52.6) |
1 (7.6) |
|
SD |
30 (34.1) |
15 (33.3) |
16 (29.6) |
1 (9.1) |
9 (47.4) |
3 (23.1) |
|
PD |
13 (14.8) |
3 (6.7) |
13 (24.1) |
3 (27.3) |
0 (0.0) |
7 (53.8) |
|
Not evaluable |
6 (6.8) |
1 (2.2) |
6 (11.1) |
1 (9.1) |
0 (0.0) |
2 (15.4) |
|
ORR (95% CI), (%) |
44.3 (34.4–57.2) |
57.8 (43.3–71.0) |
35.2 (23.8–48.6) |
54.5 (28.0–78.8) |
52.6 (31.7–72.7) |
7.6 (0.0–35.4) |
|
DCR (95% CI), (%) |
78.4 (68.6–85.8) |
91.1 (78.7–97.0) |
64.8 (51.5–76.2) |
63.6 (35.2–85.0) |
100.0 (80.2–100.0) |
30.8 (12.4–58.0) |

During osimertinib treatment follow-up (median follow-up period, 25.1 weeks), the 88 patients exhibited a median PFS time of 32.7 weeks (95% CI, 26.6–38.3 weeks), and did not reach a median OS time. Notably, the median PFS time was significantly longer (45.0 weeks) in 45 patients that exhibited T790M-positive tumor tissue samples (95% CI, 38.0–52.0 weeks) than in 13 patients who exhibited T790M-negative tissue and T790M-positive plasma samples (9.4 weeks; 95% CI, 7.2–11.0 weeks;
p<0.0001) (
Fig. 2A). Similarly, dividing the patients into tissue T790M-positive and plasma T790M-positive groups revealed a marked difference in their median PFS time, which was 45.0 and 20.4 weeks (hazard ratio=0.38;
p=0.0005), respectively (
Fig. 2B). In tissue T790M-positive patients (n=45), the median PFS was not reached, 48.1 weeks, and 20.4 weeks in T790M plasmanegative, unknown, and positive patients, respectively (
p=0.0029) (
Fig. 2C). Furthermore, plasma T790M-positive patients (n=54) who exhibited tissue samples that were T790-Munknown, -positive, and -negative displayed a median PFS time of 31.9, 20.4, and 9.4 weeks, respectively (
p=0.0065) (
Fig. 2D).
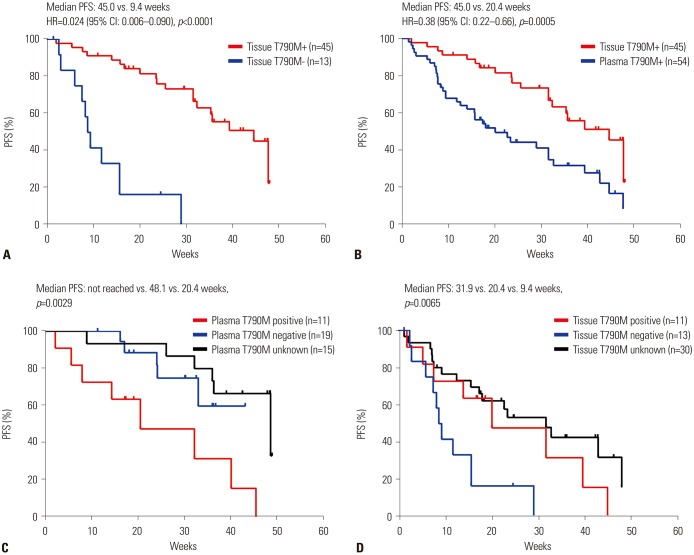 | Fig. 2PFS of patients treated with osimertinib (n=88). Survival curves of patients with documented tissue T790M mutation status (A), with tissue T790M+ versus plasma T790M+ (B), with tissue T790M+ by plasma T790M status (n=45) (C), and with plasma T790M+ by tissue T790M status (n=54) (D). CI, confidence interval; PFS, progression-free survival; HR, hazard ratio.
|
Go to :

DISCUSSION
Since the third-generation EGFR-TKI osimertinib was demonstrated to be significantly more effective than various cytotoxic agents used to treat T790M-mutant NSCLC, rebiopsy upon NSCLC progression has become an established part of standard care for these patients.
8 Clinical use of osimertinib requires confirmation of the presence of T790M mutation in tumor tissues; however, a substantial portion of patients do not currently undergo tissue rebiopsy due to a range of reasons, including poor performance status after progression, comorbidity, unavailability of next-line of therapies, inaccessible tumor locations, patient refusal, etc.
171819 In addition, the current yield of rebiopsy samples is only sufficient for genetic screening in approximately 70–80% of cases.
10202122 Therefore, non-invasive collection and screening of cell-free plasma or urine DNA has been suggested as an alternative method to overcome these limitations. Various assays with varying sensitivities and specificities have been, and continue to be, developed to screen these sample types.
23 However, to date, only the cobas
EGFR Mutation Test kit (v.2, Roche Molecular Systems, Inc., Pleasanton, CA, USA) has been approved by the FDA for plasma genotyping in the clinical setting.
24 Given the current limitations of tissue rebiopsy methods, continued development and optimization of such assays are essential to improve outcomes for patients with
EGFR-mutant NSCLC.
In the present study, we investigated the frequency, method, and success rate of tissue rebiopsy, and evaluated the incidence of T790M mutations identified in tissue and plasma samples collected from a cohort of real-world patients upon their acquisition of resistance to first or second-generation EGFR-TKIs. To the best of our knowledge, this is the largest dataset to date to prospectively analyze and compare the real-world efficacy of tissue rebiopsy and plasma genotyping.
In the current study, overall rebiopsy rate was approximately 60%, consistent with previous studies.
1021 Among 188 patients who progressed upon EGFR-TKIs, only a quarter of patients were proven to have tissue T790M
EGFR mutations (n=48), but the positive rate of T790M was 53.3% in available
EGFR mutation result (n=48/90), which was consistent with historical data.
8 The most common site of tissue rebiopsy was the lung, and the most commonly utilized rebiopsy technique was a lung biopsy via bronchoscopy (n=49, 42.2%), followed by a percutaneous lung biopsy, as previously described.
202225 Notably, pleural effusion and ascites samples, which are considered to be easily accessible sample types, were shown to be a good source of DNA for
EGFR mutation testing. The two sample types provided a sufficient sample volume for
EGFR screening in 81.8% and 100% of cases, respectively, and 100% of the collected malignant ascites fluid samples were found to harbor T790M mutation. Likewise, T790M mutations were detected in all surgical lung rebiopsy samples (n=6), suggesting that this is a promising DNA source for patients able to tolerate general anesthesia and surgery. In only one case, we identified T790M mutation in cerebrospinal fluid. Rebiopsy complication rate in the present study was negligible, comprising only a single case each of bleeding and pneumothorax resulting from bronchoscopic biopsy and CT-guided lung biopsy, respectively. Together, these findings highlight the clinical importance and advantages of identifying feasible (including non-standard) rebiopsy sites to facilitate the collection of tissue and/or body fluid samples for T790M mutation screening.
Liquid biopsies, including
EGFR genotyping assay used to screen ctDNA in patients with NSCLC in the present study, are being rapidly established as effective diagnostic tools in clinical practice. A combined analysis of the previously conducted AURA extension (
NCT01802632) and AURA2 (
NCT02094261) studies showed that the cobas
EGFR Mutation Test detected the plasma T790M mutation with PPA, NPA, and OPA levels of 61% (95% CI, 57–66%), 79% (95% CI, 70–85%), and 65% (95% CI, 61–69%) against the tissue result as reference.
26 Furthermore, a previous report showed a PNA clamp-assisted melting curve method similar to the one used in the present study to exhibit a 56.3% overall concordance rate in the detection of T790M mutation between plasma and tissue result at the time of disease progression.
27 While this previous study only analyzed a small patient cohort (n=16), our present study demonstrated a similar finding (OPA, 53.3%; 95% CI, 42.2–64.2%) using a much larger patient population (n=75). Notably, both the current and the previous study reported markedly lower NPA, OPA, and particularly PPA levels compared to those calculated for combined AURA extension/AURA2 data. This discrepancy may be a result of differences in the respective sample sizes and threshold values used by the three studies; therefore, further validation of the present findings using a larger set of paired samples is needed to confirm their clinical implications.
The secondary objective of the present study was to examine whether the clinical outcomes of osimertinib treatment in the patient cohort was correlated with the patient T790M status and/or the utilized DNA source (i.e., tissue or plasma). Oxnard, et al.
28 previously published an excellent and timely study, in which they screened ctDNA collected from patients with advanced NSCLC who received osimertinib treatment. That study reported a very similar median PFS time for tissue T790M-positive and plasma T790M-positive patients (9.7 months; 95% CI, 8.3–12.5 months, and 9.7 months; 95% CI, 8.3–11.1 months, respectively), and therefore suggested that
EGFR mutation analysis of liquid rebiopsied ctDNA may be a promising alternative method to detect T790M mutations. In contrast, the present study showed tissue and plasma T790M-positive patients to have remarkably different ORR and median PFS times (plasma, 35.2% and 20.4 weeks respectively; tissue, 57.8% and 45.0 weeks, respectively). Therefore, the efficacy of osimertinib in the present study was less predictable from the results of the plasma compared to tissue screening. This discrepancy may be a result of the fact that, unlike in the study by Oxnard, et al.,
28 a high proportion of patients in the present study who exhibited T790M-positive plasma samples (n=54) provided tissue samples that were T790M-negative (n=13, 24.1%) or -unknown (n=30, 55.6%). In fact, only 20.3% (11/54) of plasma T790M-positive cases provided T790M-positive tissue samples. The median PFS time reported by the previous and the present study for tissue T790M-negative/plasma T790M-positive patients was very similar (2.8 months and 9.4 weeks, respectively). Therefore, the seemingly inferior efficacy of osimertinib in plasma T790M-positive patients in the present study is likely reflective of the high false-positive rate (54.2%, 13/24) incurred by the utilized plasma PNA clamping-assisted fluorescence melting curve analysis method (
Table 3). Another previous study used the BEAMing technique to screen plasma samples for T790M mutation with a success rate of 13.9%.
26
The results of the present study identified a number of differences between the plasma and tissue T790M-positive patients. For example, plasma T790M-positive patients exhibited poorer baseline characteristics, and received more extensive treatment than tissue T790M-positive patients. For example, patients from these two groups predominantly received three and one line of therapy, respectively. Furthermore, 50% (27/54) of the plasma and only 24.4% (11/45) of tissue T790M-positive patients were previously treated with three or more therapy regimens. In addition, a greater proportion of plasma (74.1%, 40/54) than tissue (48.9%, 22/45) T790M-positive patients developed baseline central nervous system metastases. While the present study reported a shorter median PFS for plasma T790M-positive patients than was demonstrated by Oxnard, et al.,
28 both studies showed tissue T790M-negative patients to exhibit a markedly shorter median PFS time than T790M-positive or -unknown patients (
Fig. 2D).
Also consistent with the study by Oxnard, et al.,
28 19 of the present plasma T790M-negative cases that exhibited discordant tissue T790M-positivity experienced excellent patient outcomes, including 52.6% ORR, 100.0% DCR, and failure to reach a median PFS within the duration of follow-up period (
Fig. 2C). In fact, this PFS time was better than that experienced by plasma/tissue T790M-positive cases (median PFS time, 20.4 weeks). It may be that, in the absence of circulating T790M clones, the presence of T790M-positive tumors may indicate a more indolent form of NSCLC, which is characterized by a better prognosis and better predicted response to the administration of third-generation EGFR-TKIs.
The current study had several limitations, including the fact that all data were collected at a single center. This may have impacted the study by biasing the selection of rebiopsy sites and methods, since either or both are largely dependent on cost, facility protocols and capabilities, as well as the preferences, experience, and skill of treating physicians. In addition, since our center is a tertiary cancer center, most of the analyzed patients received extensive treatment for NSCLC, as evidenced by the fact that more than 30% were administered three or more lines of therapy prior to their participation in the ASTRIS trial. Also, the total number of the study participants was 188, but the number of tissue T790M or plasma T790M cases were just 33 and 23, respectively. Considering these small numbers, interpretation of our results should be accompanied with caution. Finally, only 51.1% of the analyzed patients exhibited NSCLC progression after treatment with a single line of EGFR-TKI therapy, and only a very limited number (n=7) of patients who were exposed to an alternative third-generation EGFR-TKI (olmutinib) were included in the study.
In conclusion, in the current real-world clinical settings, NSCLC rebiopsy upon resistance acquisition to EGFR-TKIs is increasingly important to facilitate the administration of third-generation EGFR-TKIs. The findings of the present study high-light the potential advantages of screening non-standard patient sample types, including ascites, and pleural effusion samples, for T790M mutations. The study also shows that, while their collection is less invasive, plasma samples may be insufficient to generate accurate or reliable T790M test results with currently available genotyping platforms. Therefore, continued research is needed to develop and evaluate the efficacy of novel genotyping platforms for EGFR genotyping using plasma samples.
Go to :










 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download