Clonorchis sinensis infects approximately 35 million people worldwide, causing various subclinical or clinical signs known as clonorchiasis.
12345 People are infected by ingestion of undercooked or raw freshwater fish harboring metacercariae of
C. sinensis.
678 In a historical context,
C. sinensis infection was one of the most common trematode infections in Korea, especially due to the cuisine based on raw fish, which was enjoyed by the inhabitants of the country.
91011
To reveal the genetic characteristics of
C. sinensis, researchers have attempted DNA analysis. Recently, parasitologists diagnosed
C. sinensis through DNA analysis on internal transcribed spacer (ITS),
12131415 cytochrome C oxidase subunit (CO),
510 and nicotinamide adenine dinucleotide hydrogen dehydrogenase (NAD) subunits.
512 The molecular analyses claimed that
C. sinensis is genetically distinct from other trematodes.
11213141516
Meanwhile, paleoparasitologists have also tried to reveal the genetic characteristics of ancient
C. sinensis through research on the samples collected at archeological sites. One of such studies was carried out in Korea. Shin et al.
17 successfully analyzed ancient DNA (aDNA) sequences of
C. sinensis eggs collected from the 17th century Korean mummy feces. They showed that amplified sequences of
C. sinensis ITS1, ITS2 and CO1 were completely identical to modern
C. sinensis sequences in GenBank.
17
Although this pioneering work was to reveal genetic traits of ancient
C. sinensis, the number of aDNA cases reported so far was too insufficient to get detailed information of ancient
C. sinensis genetics. Fortunately, by paleoparasitological studies in Korea over the past several years, we collected a number of pre-modern Korean mummy feces or tissue specimens in which the presence of ancient
Clonorchis eggs was microscopically confirmed.
1819 Utilizing the ancient specimens, in this study, we analyzed CO1, ITS1, NAD2 and NAD5 of
C. sinensis aDNA. The current report could expand the spatiotemporal scope of parasitological research about the genetic history of
C. sinensis.
The samples used in this study were obtained from the 16th to 17th century Joseon mummies (n = 5; Andong, Cheongdo, Dalsung, Hadong1 and Mungyeong) (
Table 1,
Supplementary Figs. 1 and
2). The specimens were coprolites retrieved from mummy intestines (Andong, Dalsung, and Hadong1) or mummified livers (Cheongdo and Mungyeong). We followed the
Criteria of Authentication for authentic aDNA analysis.
20
For aDNA extraction, we followed the method in our previous report.
21 The specimens (0.3 g) were treated in a lysis buffer (1 mL) for 24 hours at 56°C. DNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1) and then chloroform/isoamyl alcohol (24:1). DNA isolation/purification was performed by a QIAmp PCR purification kit (Qiagen, Hilden, Germany). Extract DNA (10 μL) was treated with 1 unit of uracil-DNA-glycosylase (New England Biolabs, Ipswich, MA, USA) for 30 minutes at 37°C. It (40 ng) was then mixed with a reagent premix containing 10 pmol of each primer (
Table 2) and 1X AmpliTaq Gold® 360 Master Mix (Life Technologies, Camarillo, CA, USA). PCR conditions were as follows: pre-denaturation at 95°C for 10 minutes; 45 cycles of denaturation at 95°C for 30 seconds, annealing at 54°C–63°C for 30 seconds, extension at 72°C for 30 seconds, and final extension at 72°C for 10 minutes. The amplified PCR products separated on 2.5% agarose gel (Invitrogen, Waltham, MA, USA) were stained by ethidium bromide. Electrophoresis also included negative (extraction) controls.
The PCR amplicon was isolated by a QIAquick Gel Extraction Kit (Qiagen). Bacterial transformation was done using a pGEM-T Easy Vector system (Promega Corporation, Madison, WI, USA). Transformed bacteria were then grown on agar plate containing X-GAL (40 μg/μL), ampicillin (50 μg/mL) and 0.5 mM IPTG for 14 hours. After colonies were grown in LB media for 12 hours, the cultured bacteria were purified by a QIAprep® Spin Miniprep kit (Qiagen). Each amplified DNA strand was sequenced by an ABI Prism BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Waltham, MA, USA) and 3730xl Automatic Sequencer (Applied Biosystems).
To obtain consensus sequence, multiple sequence alignment was performed for each aDNA region by Clustal W implemented in MEGA7.
22 We compared the consensus sequences of ancient
C. sinensis to GenBank taxa by NCBI/BLAST tools.
23 The evolutionary relationship of ancient
C. sinensis and other parasites of NCBI GenBank was inferred by the
Phylogeny Reconstruction analysis implemented in MEGA7. We used Maximum Likelihood method. Selected parameters are Tamura 3-parameter
24 (CO1), Kimura 2-parameter
25 (ITS1), Hasegawa-Kishino-Yano model
26 (NAD2 and 5) for Model/Method. We performed bootstrap test to estimate the reliability of the tree. The number of bootstrap replicates was 500.
27
To select the specimens used for aDNA analysis, we screened all the mummy coprolite samples using PCR with
C. sinensis primers for CO1 (206 bp), ITS1-2 (122 bp), NAD2-1 (194 bp) and NAD5-1 (164 bp). In agarose gel electrophoresis, negative (extraction) controls exhibited no amplified bands. In Andong feces, the PCR products were detected for
C. sinensis CO1, ITS1-2, NAD2-1 and NAD5-1. Mungyeong specimen also showed positive results for
C. sinensis CO1 and ITS1-2 (
Supplementary Fig. 3). We thus used the Andong and Mungyeong specimens for subsequent aDNA analysis.
To get the consensus aDNA sequences of
C. sinensis CO1, ITS1, NAD2 and NAD5, we tried to do cloning and sequencing of each specific amplicon. By these trials, 9–10 clone sequences were successfully acquired for CO1, ITS1, NAD2 and NAD5 amplicons (
Supplementary Fig. 4). The total sizes of consensus sequences obtained by multiple sequence alignment were 162 bp (CO1), 431 bp (ITS1), 588 bp (NAD2) and 443 bp (NAD5), respectively. The
C. sinensis consensus sequences of Andong and Mungyeong specimens were almost the same to each other, except for a little difference at a nucleotide position (transversions occurred in the positions CO1: 100 and ITS1: 167) (
Fig. 1). Considering these results, we conjecture that genetic characteristics of ancient
C. sinensis might not have been uniform during the Joseon period.
In BLAST searching, the
C. sinensis consensus sequences of Andong and Mungyeong specimens were completely or almost identical to
C. sinensis CO1, ITS1, NAD2 and NAD 5 sequences reported in GenBank (
Table 3 and
Fig. 1). Briefly, the current ancient
C. sinensis CO1 sequences were 100% identical to GenBank sequences of
C. sinensis reported from Korea (KY564177.1), China (FJ965391.1; FJ965383.1; AF188122.2; AF184619.2), Russia (MF406205.1; MF406204.1), and Vietnam (MF287785.1; KJ204609.1).
C. sinensis ITS1 sequences of Korean mummies also exhibited very high similarities (99%) to the GenBank ITS1 sequences reported from Korea (JN638318.1; JN638320.1), China (KJ137226.1; KF740425.1; HQ186255), Russia (JQ048578.1; KC987517.1) and Vietnam (MF319655.1; MF319650.1; MF319653.1). The aDNA sequences of Andong and Mungyeong specimens were also completely or almost (99%) identical to GenBank
C. sinensis NAD2 and NAD sequences from Korea (NAD2, JF729304.1; NAD5, FJ729304.1), China (NAD2, KC170192.1; NAD5, KY564177.1), Russia (NAD2, FJ381664.2; NAD5, FJ381664.1) and Vietnam (NAD2, AY264851.1) (
Table 3 and
Fig. 1).
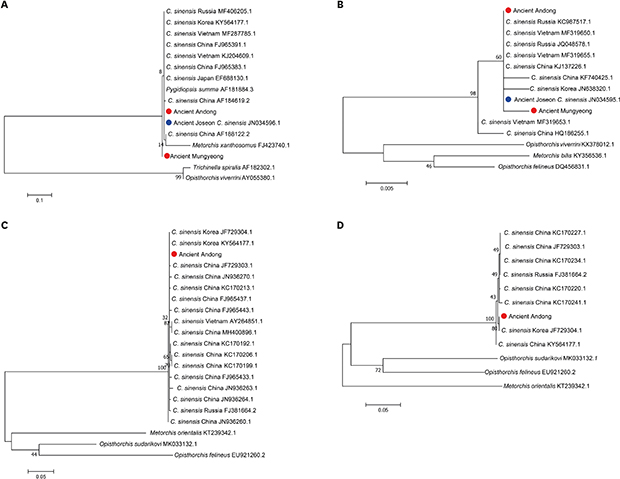
In the analyses, we found that CO1 region could not be an effective marker for differential diagnosis between
C. sinensis and other trematode species because the CO1 sequences of
Pygidiopsis summa (AF184884.3) and
Trichinella spiralis (AF182302.1) were not distinguishable from
C. sinensis CO1 sequences. Meanwhile,
C. sinensis ITS1, NAD2 and NAD5 sequences were clearly distinct from those of other trematode species (
Fig. 1). We identified similar patterns in phylogenetic analyses (
Fig. 2). In case of CO1, ancient Andong and Mungyeong sequences belonged to the clade not only with
C. sinensis, but also with
P. summa and
M. xanthosomus. On the other hand, ITS1, NAD2 and NAD5 of
C. sinensis and other trematode species were separately clustered into different clades (
Fig. 2). Actually, previous studies proposed that the interspecific sequence variations within zoonotic trematodes were observed for ITS1, NAD2 and NAD5.
13 In this study, we re-confirmed the usefulness of ITS1, NAD2 and NAD5 as molecular markers for differential diagnosis of
C. sinensis from other trematode species.
In summary, our present study about C. sinensis aDNA retrieved from Korean mummies is designed to uncover invaluable genetic information of C. sinensis prevalent among pre-20th century Korean people. Although detailed understanding of C. sinensis genetics require a future retrieval of ancient or modern DNA sequences in wider geo-historical scope, our current report represent a significant step to improve our knowledge about genetic history of C. sinensis.









 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download