INTRODUCTION
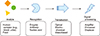
A biosensor is a device that enables the detection and analysis of biomolecules. The sensor consists of recognition, transduction, and signal processing components. When a sample is applied to the device, the sensor first identifies the target analytes, and then the transducer transforms the recognition into measurable signals for the detector to process.1 The overall concept is summarized in Fig. 1. The most common biosensor is the glucose meter, which measures blood glucose levels by utilizing an enzyme called glucose oxidase (GOx), converting glucose in the blood to gluconic acid and hydrogen peroxide.23 Then, the electrochemical transducer can sense the number of electrons induced by the deoxidization of H2O2 during the reaction and transform it into measurable signal for the amplifier to process. The read out is thus based on the electron flow which is proportional to the concentration of glucose molecules existing in the blood. This review will briefly introduce each sensor component, and then discuss specific examples about nucleic and exosome detection.
MAJOR COMPONENTS ASSOCIATED WITH BIOSENSOR
1. Recognition
The bio-recognition component, also known as a bioreceptor, specifically recognizes target analytes (e.g., binding), leading to signal generation that is used by the transducer to enable detection.4 Common receptors for biosensors include enzymes, antibodies, and nucleic acids. Table 1 summarizes the advantages and drawbacks of various recognition elements for biosensors.
1) Recognition moieties
Enzymatic bioreceptors catalyze reactions, converting analytes into a detectable signal. The receptor specifically recognizes the target analytes, and is reusable for multiple detections over time since enzymes are not consumed during the reaction. For example, Palygin's group developed two sensor designs with enzyme recognition components to quantify endogenous ATP and H2O2 in the kidneys.5 Both designs involve the sequential catalytic reactions of glycerol kinase and glycerol-3-phosphate oxidase, locating in the enzymatic layer of the sensor and are activated by the presence of ATP. Due to the enzyme stability, these sensors potentially exhibit more limited lifetimes.6
Antibody recognition is mainly utilized for immunosensor. the antibody, also known as immunoglobulin, has a Y shape conformation with paratopes on each tip that can specifically recognize the epitopes on antigen.7 Therefore, the sensor can utilize the binding to enable the antigens' concentration quantification via the direct signal output to transducer or by the indirect reaction of competitive and displacement.8 Kaushik's group recently developed an immunosensing chip for diagnosing Zika-virus infection.910 The device has ZIKV-envelop protein antibody immobilized onto the self-assembled monolayer to capture the Zika virus specifically with high affinity, resulting in the capability of quantifying virus concentration. This sensor exhibits potential for use in rapid disease diagnosis and may be applied in the point-of-care domain.10 However, a potential drawback is antibody stability11 and the cost associated with antibody is also a concern.12
Biosensors utilizing nucleic acids as the recognition component, are categorized as affinity sensors. The sensor detects the binding reaction and the associated physicochemical changes between the nucleic acid and analyte, allowing nucleic acid-based biosensor to be commonly used in detecting a wide range of targets like bacteria and viruses.13 However, since the nucleic acid is prone to being affected by the environmental condition like temperature and pH and is incapable of resisting being cleaved by the restriction enzyme, some of the utilities have been more limited.14 Among all the biosensors that use nucleic acids, aptamer-based ones have been drawing lots of attention owing to their high thermal stability and low immunogenicity.15 Aptamer is an oligonucleotide sequence selected from a large pool that can be engineered to bind with specific target biomolecule with high affinity.16 The single strand nucleic acid can bind through electrostatic interactions, hydrophobic interactions, or their complementary shapes.17 Aptamers can be developed to bind with almost any target, from small molecules to microorganisms, giving it a wild variety of applications. Aptamer is also good for multiplexing with the high-density immobilization due to its small size.18 Dalirirad et al.19 utilize aptamer immobilized on the gold colloid surfaces as the recognition component for sensing cortisol. When the cortisol molecules go through the lateral flow strip assay, they will react with the aptamers and result in desorption from the gold surface. The free gold nanoparticles will then go on and react with the cysteamine at the test zone and be captured, which produces visual signal.19
2) Surface functionalization
The surface modification enables target molecules binding only to the specific recognition region on the sensor;20 in the meantime, the surrounding area should be kept inert to ensure minimum sample consumption.21 The surface functionalization includes anchors, spacers and recognition moiety.22 Anchors are the molecular interactions between the receptors and sensing surfaces such as a chemical bond. Weak anchoring involves no chemical bond formation during the process, also known as physisorption. Since the weakly adhering anchor might have a residence time on interface shorter than it requires for the recognition component to function, multiple anchor units will have to be linked together for stable attachment on the surface. An approach to circumvent this issue is to utilize multivalent anchors, meaning multiple physical adsorptions between the anchor and surface occurs simultaneously.23 Mejri et al.,24 makes use of the fast trait of physisorption to immobilized antibody on the substrates. This method performs better on the detection of bacteria than other more complicated antibody immobilization approaches. On the other hand, strong anchors, also referred to as chemisorption, immobilize a molecular over a long period of time on the surface through electron-sharing interaction. Liu et al.25 utilize the chemisorption surface modification method for a novel graphene-based biosensor uses. The highly efficient enzyme electrode was directly obtained by covalently linking the carboxyl groups of graphene oxide sheets to amines of glucose oxidase. This fabrication technique possesses reproducibility and good storage stability. Spacers can improve recognition efficiency by placing the receptor further into the sample to promote interaction with analytes. The spacer can also prevent non-specific binding.26 The common approaches to form the spacer film can be categorized into two ways: grafting-to and grafting-from. Grafting-to is when the spacer and recognition moiety are pre-assembled as one single molecule, and then immobilized on the surface. Since the spacer will be chemically linked to the anchor first, the spatial pattering of the anchor on the surface can be passed on directly to the spacer and the bioreceptor, giving the opportunity to design the distribution of the recognition moiety on the surface. Grafting-from, on the contrary, immobilizes the initiators on the surface first, and then the spacer is grown via a surface-initiated polymerization reaction. This approach can produce large area films.27
2. Transducers for biosensor
The transducer is a component that converts the biochemical signals induced by recognition to measurable signals.28 The comparison of different transducers is summarized in Table 2. Optical sensors are best known for their high sensitivity and the ability to monitor from a remote location,29 but the instruments can be costly to install and are susceptible to external physical damage.30 As for the electrochemical sensor, good resolution, excellent repeatability and accuracy all make it a good approach for detection. However, they have the weakness of being easily influenced by temperature change, and the application is limited by having a short shelf-life before needing to be replaced.31 Quartz crystal microbalance (QCM) utilizes a mass-based transducer, which detects very small changes in mass and are very useful for analytes that possess no electrically conducting properties nor the florescent signals (e.g., virus). Thinner quartz sheets can increase QCM frequency for better sensitivity, but the device will be mechanically unstable and fragile.32
1) Optical transducers
Optical biosensors work on the interaction of optical fields with their biorecognition components to achieve their detection purpose.33 They are one of the most commonly used biosensors because of their sensitivity, specificity, and small footprints.34 The optical biosensors can generally be divided into two categories: label-free and label-based. Label-free sensing means that the signal is directly generated from the interaction between the sample and the transducer. In the label-based approach, the target analyte has to be tagged with a reporter molecule, which enables the detection via fluorescent, luminescent, or colorimetric signals.35 Commonly seen optical biosensors includes, fluorescent, surface plasmon resonance (SPR), interferometer, optical waveguild, and ring resonator. The SPR phenomenon occurs at the interface of the two conducting materials (e.g., glass and metal) when polarized light illuminates at a specific angle.36 This will generate a surface plasma wave along the surface. Since the wave is on the boundary of the conductor and the external medium (air, water or vacuum for example), these oscillations are very sensitive to any change of this boundary, such as the absorption of molecules to the conducting surface. By measuring the changes in wavelengths, reflectivity, and angles against time, we can attain the detection result.37 SPR sensors such as Biacore™ are used in the measuring of antibody-antigen interactions like binding affinities, kinetic rate constants and thermodynamics. The recognition component is immobilized on the sensor chip surface and followed by the injection of the analyte flowing across the surface. The changes in the index of refraction at the surface caused by binding interactions are detected and recorded as resonance units by the sensor.38 Another type of optical transducer is the ring resonator. When binding with the surface, the absorption of the molecules increases the refractive index, which alters the overall effective index and leads to a shift in the cavity's resonance wavelength. The difference can be determined through a tunable laser source.39 Flueckiger et al.40 have developed a sub-wavelength gratings (SWG) ring resonator designed for transverse electric polarized light to enable more sensitive detection over the transverse magnetic ring resonator. The SWG waveguide can minimize the mismatch loss and improve the field overlap of biomolecules on the waveguide's surface by engineering the effective refractive index, which is the ratio of the propagation constant for a given polarization in the direction of propagation in a waveguide structure to the free space propagation constant.
2) Electrochemical transducer
Electrochemical biosensors convert analyte recognition to an electronic signal, which exhibits advantages such as simplicity, lower cost, excellent detection limits, and robustness. It also has the ability to analyze small amounts of samples and the output is easy to read-out and process. Electrochemical biosensors mainly use enzymes as the recognition moiety (one of the exceptions is immunosensor utilizing antibody-antigen) because of the enzyme biocatalytic activity and specificity.41 Electrochemical transducers can be divided into three main categories.42 First, amperometry, which measures the current caused by the electrochemical oxidation or reduction of an electroactive species. The resulting current is proportional to the analyte concentration. The second one is potentiometry, which measures the potential between two different electrodes when there are no significant current flows between them using a high impedance voltmeter. Potentiometry can present the information about ion activities in electrochemical reactions. Lastly, the conductometry, which measures the analyte's ability to conduct an electrical current between electrodes or reference nodes induced by biorecognition events that change ionic species' concentration.41 For example, Soldatkin et al.43 developed a sensor that is highly sensitive to substrate (sucrose) and heavy metal ions, especially to Hg2+ and Ag+, using immobilized enzymes (e.g., invertase, mutarotase, glucose oxidase) to generate signals, caused by the enzymatic reaction inhibition via changes of the medium conductivity.
3) Mass-based transducer
The mass-based biosensors, also known as gravimetric biosensors, apply the basic principle of a response to a change in mass.44 Most gravimetric biosensors use piezoelectric quartz crystals, which can either be in the form of resonating crystals (quartz crystal microbalance, QCM), or as surface acoustic wave (SAW) devices. Since these have ease of use, shorter analysis time, low cost, and the ability to produce label-free measurement, QCM based sensors have been a star in the area of rapid detection of pathogens45 and toxins46 for the past few years. Typically, a QCM biosensor consists of an AT-cut quartz crystal wafer in the middle of two metal electrodes, and the resonant frequency of QCM will change according to the mass change at the crystal's surface. The decrease in frequency is proportional to the mass on the sensor. As a result, by combining the QCM devise with highly specific interaction such as antigen-antibody it can enable the direct detection of micro-organisms.47 SAW based biosensors can detect acoustic waves generated by mass loading on the surface of the piezoelectric crystal via the interdigital transducers (IDT).48 The IDT allows the acoustic energy to be strongly confined to the surface no matter the thickness of the substrate. The analyte recognition by the immobilized receptors changes the velocity of SAW and produces signal by the driving electronics.49
3. Signal processing
Since the signal generated by the transducer is usually weak and layered with relatively high noise backgrounds, it is important to process the signal before the result can be analyzed.50 The signal processing part of a biosensing device is an electronic system which includes a signal amplifier, processor, and a display. The signal collected from the transducer is intensified, and then converted to a digital form before passing to a microprocessor stage where the data is processed and ready for downstream analyzing. The processor usually subtracts the raw collected signal with a reference baseline, derived from the matching transducer without analytes, to filter out the unwanted background noise.51
APPLICATIONS OF BIOSENSORS
Currently, biosensor technology is being applied in diverse fields such as clinical monitoring,252 in vitro diagnostics,53 and the food industry.54 In addition to common glucose monitoring,55 biosensors have great potential in diagnosing cancer. For example, Atay et al.53 have developed QCM-based biosensors to detect highly metastatic breast cancer cells. Biosensors are also frequently used in analyzing food security, for instance, the detection of compound contaminants, allergens, toxins, pathogens, additives, etc., during quality control processes.54 This review will focus on the biosensors for detecting nucleic acid targets and exosomes.
1. Biosensors for nucleic acid detection
As we are advancing in genomics research, nucleic acid detection is attracting more and more attention. The development of biosensors for nucleic acid detection has many benefits related to biochemical studies,56 medical diagnosis,57 and therapeutic applications.58 For example, Guo et al.58 reported a surface enhanced raman spectroscopy (SERS) nanosensor which can simultaneously detect multiple disease-related nucleic acids for early disease screening, such as prostate carcinoma, and helping doctors make treatment decisions. Wang et al.57 developed a catalytic hairpin assembly-based (CHA) electrochemical biosensor for detecting the liver cancer related short gene MXR7. In food research, Xu et al.56 reported using a nucleic acid biosensor which combines loop-mediated isothermal amplifications for universal mammalian identification, especially meat adulteration on site. The biosensors for nucleic acid detection usually utilize hybridization of nucleotide probes and target sequences for recognition with various transducing mechanisms, including optical, electrochemical, and mass-based biosensors.59
1) Optical biosensors for nucleic acid detection
(1) SPR-based biosensors for nucleic acid detection
SPR biosensors are perhaps some of the most widely-used optical biosensors, based on a physical phenomenon called surface plasmon resonance.60 When SPR is applied in sensors, the recognition events interfere with the surface plasmon wave, leading to a reduction of the reflected light intensity that is proportionate to the mass change from recognized molecules on the surface. In order to utilize this phenomenon for biosensing, recognition moieties are immobilized on surface of a metal, such as gold. When the target molecules flow through the sensing surface, the target molecules are captured and influence the surface plasmon wave. The resulting effect is later, at the presence of incident light, displayed as a change of reflected light intensity on the detector.
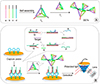
Diao et al.61 developed a SPR-based biosensor for HIV-related DNA detection by combining entropy-driven displacement reactions (ESDRs) and double layer DNA tetrahedrons (DDTs). DDTs (Fig. 2A) are dendritic DNA tetrahedron, which is a macro biomolecule with excellent mechanical rigidity and structural stability.62 ESDRs, also known as toehold-mediated strand displacement, are an isothermal and efficient signal amplification technique in which the reaction is driven forward exclusively by entropy force.61 The ESDRs need delicately-designed sequences. In this study, three-stranded DNA complexes (Q, P, and R) in Fig. 2B form the initial DNA complex. The target sequences (T) interacts with the initial DNA complexes to release P sequences. Then, the resulting complexes interact with F sequences, leading to FQ double-stranded DNA complex formation by releasing T and R sequences. The amplification is achieved by continuously producing FQ double-stranded DNA complexes and the simultaneously-released T sequences can enter the next cycle to further interact with other available three-stranded DNA complexes. Once the FQ double-stranded DNA complexes are recognized by the capture probes on the surface (Fig. 2B). The DDTs will label the captured FQ double-stranded DNA via hybridization to increase the complex molecular weight for stronger SPR signaling. This approach can detect target DNA in a linear range from 1 pM to 150 nM with a detection limit of 48 fM within 60 minutes.61
(2) LSPR-based biosensors for nucleic acid detection
Compared to SPR, an optical phenomenon on large metal structures, localized SPR (LSPR) is a phenomenon that occurs on metallic nanostructures.63 When an incident light is introduced to the metallic nanostructure, the electromagnetic field of the light induces collective electron charge oscillations confined in the metallic nanostructure, and consequently leads to an absorbance of light within the ultraviolet-visible (UV-VIS) band. Therefore, the biorecognition events in LSPR sensors are observed by measuring the absorbance wavelength shift. Due to the nanoscale metal and special characteristic described above, LSPR sensors avoid the complex instrument settings required by traditional SPR sensors and provide researchers with a simpler and cheaper alternative for biomolecule sensing.
Klinghammer et al.63 developed a microfluidic-channel-integrated LSPR biosensor, utilizing large arrays of gold nanorods. The sensor demonstrates the real-time and label-free detection of short oligonucleotide sequences in the low ng/µL range, intending to support water, food, and drug screening in a cost-effective way. In this work, probe DNA (25 bp), immobilized on the gold nanorod surfaces, is complimentary to two cDNA targets, 25 and 100 bp, with identical 25 bp sequences. cDNA recognitions on the surface induce the signal for the LSPR sensor. The data indicates that the sensor exhibits a limit of detection around 250 nM for both 25 and 100 bp cDNA molecules.63
(3) Fluorescence-based biosensors for nucleic acid detection
The fluorescent biosensor is another widely-used optical biosensor, which detects the frequency of change of the electromagnetic emissions after a fluorescent dye is exposed to radiation. Other than directly detecting fluorescent labels (e.g., fluorescein) associated with the target molecules, the sensor can also employ fluorescence resonance energy transfer (FRET) for detection. FRET is a phenomenon when two fluorophores are in close vicinity, where exiting donor fluorophore leads to the emission of an acceptor fluorophore, because the donor emission wavelength overlaps with the acceptor excitation wavelength. Two common ways to incorporate FRET into biosensors are: (a) incorporate both the donor and acceptor on the recognition moiety in such a way that they are far enough and cannot undergo FRET and binding with target molecules causes the conformational change of the recognition moiety, bringing the pair fluorophores in proximity for FRET, or (b) the donor and acceptor are incorporated on the target molecule and the recognition moiety respectively, where the binding event brings the two fluorophores close enough to undergo FRET. Further, depending on diverse experimental settings, there are many strategies other than FRET.6465
Zheng et al.66 developed a fluorescence-based biosensor for detecting thrombin and adenosine, using a fluorescent-labeled hairpin on the surface of gold nanoparticles. In the design, one end of the hairpin is grafted to the gold surface, and the other end is labeled with a fluorophore. The immobilized hairpin structure positions the fluorophore very close to the gold surface, resulting in a quenched state. At the presence of target sequences, the hairpin structure opens via hybridization, which moves the fluorophore away from the gold surface, switching to fluorescence. Then, another hairpin in the solution is utilized to release the hybridized target sequence by forming a stable double strand DNA with the hairpin on the surface, keeping the fluorophore away from surface. The released target sequence can subsequently trigger the second round of hairpin opening events, resulting in an amplified fluorescent signal. In this study, the detection limit of the sensor for target nucleic acids is estimated to be as low as 3.4 pM with a linear detection range from 15 pM to 370 pM, and the optimum reaction time is 3 hours.66
2) Electrochemical biosensors for nucleic acid detection
Electrochemical biosensors are based on detecting electric property changes such as electric current or potential.67 These changes are often caused by chemical reactions in which the analytes directly participate in generating or absorbing electrons or indirectly trigger a change of electron amounts in solution. Potentiometric biosensors and amperometric biosensors are the two main categories. Potentiometric biosensors detect electrical potential changes between the ion-selected electrode and reference electrode.68 The potential change results from the accumulated ions on the working electrode, typically caused by enzymatic reactions. Amperometric biosensors detect the electrical current generated from the redox reaction of electroactive analytes on the electrode surface being detected under controlled potential conditions.67 Therefore, the well-known voltammetric biosensor is also considered to be an amperometric biosensor.67 Voltammetric biosensors detect target analytes by measuring the current induced under various potentials, including differential pulses, polarography, linear sweeps, differential staircases, normal pulses, reverse pulses, cyclic voltammetry, etc. Electrochemical biosensors such as glucose meters have had significant development in recent years because they are sensitive, fast, low-cost, and easy to operate.67 Many strategies have been developed to utilize electrochemical properties to detect bio-markers such as nucleic acids,69 proteins,70 bacteria,71 viruses,72 extracellular vesicles,73 and amino acids.74
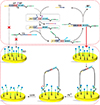
Wang et al.75 report an amperometric biosensor utilizing a two-leg DNA walker for amplified electrochemical detection of nucleic acids, which can potentially be employed for POC applications such as diagnosing gene-related diseases. The amplified electrochemical signal is generated due to a target DNA-triggered cyclic reaction.75 In addition to the target DNA (TD), this strategy includes template strands (TS), fuel strands (FS), assist probes (AP), substrate strands (SD), and block probes (BP), as shown in Fig. 3. The sensor detects the signal generated via the electron transfer between methylene blue (MB), attached to one end of the SD, and the gold electrode. FS and TS are engineered with metal-dependent DNAzyme tail sequences.75 Initially AP and BP are partially and fully hybridized on the same TD to form a three-stranded complex. TD initiates the hybridization with the complex from the TD toehold, which causes the release of AP. The resulting complex exhibits a single strand DNA sequence in the middle of TD that is complementary to FS. Then, the complex hybridization with FS releases TD and BP by branch migration. The resulting molecule is the two-leg DNA walker because there are two protruding DNAzyme tails at either end. At the same time the released TD starts the next reaction to form more DNA walkers. The gold electrode surface is pretreated with SD that is partially complementary to the sequence of the DNAzyme on DNA walker and SD is end-labeled with MB. After the addition of Pb2+, the two-leg DNA walkers catalyze the cleavage of SD with MB. The cleavage does not occur without TD, so the MB group is holding close to the gold electrode to enable electron transfer. The appearance of TD triggers the cyclic reaction and generates large amounts of DNA walkers to cleave the SD with the MB groups, which consequently interrupts the electron transfer between the MB group and the gold electrode. As a result, the amount of TD presented is proportional to the electric current reduction. In this study, a good linear relationship was observed between the TD and the ratio of (I0−I)/I0 (I0 and I represent the peak current in the absence and presence of target DNA). The sensor dynamic range is from 0.3 to 500 fM with a detection limit at 0.29 fM.75
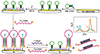
Xiong et al.76 developed an alternative strategy using an amperometric sensor with a methylene blue (MB) tag for nucleic acid detection. In this research, the human immunodeficiency virus type 1 (HIV-1) gene was chosen as the model analyte to test the performance. As shown in Fig. 4, methylene blue (MB) group is labeled at one end of a hairpin which is immobilized on gold surface via Au-S bond. After the immobilization of MB-DNA, the surface was passivated with 6-mercaptohexanol. Then, a different single strand hairpin sequence (Fc-DNA), on which two ferrocenes (displayed as orange balls in Fig. 4) were used for modification to enhance the oxidation signal, opening the immobilized MB-DNA hairpin, forming a triple-helix conformation. The addition of target DNA could hybridize with the Fc-DNA, causing disassociation from MB-DNA. Therefore, MB-DNA was restored to the original hairpin structure, resulting in the increase of electrical current. The sensor dynamic range is from 0.5 to 80 pM with the limit of detection as low as 0.12 pM.76
Zhao et al.77 report a nanopore electrochemical biosensor in which a hybridization chain reaction (HCR) strategy is applied for amplification. The biosensor aims at detecting low abundances of survivin mRNA for the early diagnosis of cancer. In this work, surviving mRNA is chosen as a model target due to its overexpression in many cancer cells. The principle of nanopore-based sensing is straightforward, the target molecules can access in or attach to the surface of a pore, thereby leading the ionic current changes to become detectable. The nanopore membrane is usually located between two electrochemical chambers that are filled with conducting buffers. Under an applied voltage, electrolyte ions flow through the nanopores, which are measured as current in the electrical instrument.78 When the target analyte appears, it will stay in the nanopore or pass through nanopore, and consequently interfere with the ion current. Furthermore, the detection of ion changes provides an analysis of the target analyte. If the target molecule passes through the nanopore, it will lead to a pulse of current change, since the ion current is back to normal after the target molecule passes through. While DNA sequencing is the common application, nanopores are also used for DNA detection. In this case, the capture of target sequence results in blocking the nanopore, causing a permanent current decrease. The nanopore biosensor achieves a sensitivity of 30 fM with a linear dynamic range from 0.1 to 10 pM.77
3) Other biosensors for detecting nucleic acid
In addition to optical and electrochemical sensors, biosensors for nucleic acid detection include mass-based biosensors, or gravimetric biosensors. Mass-based biosensors are mainly surface acoustic wave (SAW) biosensors,67 microcantilever-based (MCL) biosensors,79 and quartz crystal microbalances (QCM).80 The basic SAW sensor consists of a piezoelectric substrate with an input interdigitated transducer (IDT) on one side of the substrate and an output IDT transducer on the other side. The input IDT first transduces an input electrical signal into a mechanical wave, which unlike an electrical signal, can be easily influenced by physical phenomena. The output IDT then transduces this wave back into an electrical signal. Changes in amplitude, phase, frequency, or time-delay between the input and output electrical signals are related to the biorecognition events on the surface, typically the mass changes resulted from the target analyte, and thus can be used to detect target analytes. A microcantilever (MCL) is a simple mechanical structure, which is clamped to one end and free at the other end. When the target analyte is captured by the sensing layer on one side of the microcantilever, the mechanical stress from analytes can cause the microcantilever to bend.79 As a result, the analytical signal of MCL sensor comes from the resonance frequency and amplitude changes caused by the binding of target analyte on microcantilever. The QCM sensor utilizes an oscillator circuit containing a thin quartz disc with circular electrodes on both sides of the quartz. A varying voltage applied on the electrode can cause a mechanical oscillation of the crystal, whose frequency can be altered proportionally by the change of mass on the surface.80 Compared to the optical and electrochemical sensing techniques, these techniques are less common in nucleic acid detection.
4) Discussion for nucleic acid detection
The comparison of different biosensors for nucleic acid detection is summarized in Table 3. The SPR biosensor60 is easy to operate, stable, fast, and inexpensive, however, the strands involved in ESDRs require precise length to achieve high performance. Specifically, the Q strand toehold length can affect the signal-to-noise ratio significantly. Also, temperature is critical to nucleic acid hybridization. The data suggest that the assay is not performing ideally if the temperature is lower than 37℃. The merits of LSPR sensor62 include the throughput and multiplexing capability. Utilizing nanoantenna arrays in microfluidic system makes the sensor easy to use. However, the array density can directly impact the DNA hybridization efficiency. The sensor exhibits higher detection limits, which are partially caused by the laminar flow profile.62 The fluorescence-based biosensor65 is low-cost and exhibits sufficient sensitivity and higher versatility. Compared to the label-free sensors, using fluorophores and quenchers always raises concerns of labeling efficiency and label stability under different environmental conditions (e.g., pH). The amperometric biosensor75 shows outstanding detection limits over other strategies because of the toehold-mediated strand displacement. The other amperometric biosensor76 with a triple-helix molecular switch exhibits excellent stability, sensitivity, and selectivity, as well as a lower LOD. Moreover, the biosensor can regenerate readily with an alkaline buffer containing Mg2+ without a heat-annealing treatment. The nanopore biosensor77 shows an excellent detection limit with a wide linear dynamic range. However, the sensor exhibits low regeneration efficiency (40%–60%) and is potentially expensive.
2. Biosensors for exosome detection
Exosome, a type of extracellular vesicle, can facilitate intercellular communication in diverse cellular processes.81 Exosomes come from endocytic pathways and exhibit a diameter between 40 to 100 nm.81 In human, exosomes are involved in many cancers' development and metastases.82 The cargo of the exosomes, especially proteins, are considered as potential biomarkers for cancer screening, diagnosis, and monitoring in many preliminary studies.818283 Herein, we will briefly introduce few recent studies in which various sensing and transducing techniques were used to construct exosome biosensors.
1) Optical biosensors for exosome detection
(1) SERS-based biosensor
Surface enhanced Ramen scattering (SERS) is a label-free optical technique for biosensing.84 The approach can enhance Ramen scattering by molecules absorbed on a rough metal surface85 or by nanostructure such as plasmonic-magnetic silica nanotubes,86 with an enhancement factor reaching 1011 to detect single molecules.87
Tian et al.88 reported a simple strategy to detect exosomes by using SERS nanoprobes. In the study, exosomes derived from the hepatocellular carcinoma cell line (HepG2) are model analytes for liver cancer diagnosis. Gold nanostars modified with a bivalent cholesterol-labeled DNA anchor were used as the SERS probe (Fig. 5A). The assay started by capturing the target exosomes via immuno-magnetic beads, and then labeling the captured exosomes with the SERS nanoprobe via hydrophobic interaction between the cholesterol and lipid membranes, resulting in a sandwich complex (Fig. 5B). The resulting immunoxomplexes could be captured magnetically and deposited on a silica slide for detection. In this study, the sensor reached a limit of detection of 27 particles/µL with a linear relation between exosome concentration and corresponding SERS signal ranging from 40 to 4×107 particles/µL.88
(2) PIA-based biosensor
Plasmonic interferometer array (PIA) is an emerging technique being utilized in biosensing.89 Typically, a ring-hole plasmonic interferometer is comprised of two nano groove rings and a central slit upon a thin gold or silver film. The light illuminating the structure strikes the grooves to excite surface plasmon polariton, which interferes with the light passing through the central slit. The transmitted light is influenced by the illumination wavelength, refractive index of medium and the groove-slit distance. Capturing target analytes via the recognition moieties on the surface changes the refractive index between the central slit and grooves, resulting in the intensity change of transmitted light.89
Zeng et al.89 reported a PIA biosensor which detects EGFR+ exosomes sensitively in real-time.89 The sensor can utilize a miniaturized microscope or couple with a smart phone to enable portable and sensitive detection for early cancer diagnosis. In this study, EGFR were used as lung cancer biomarkers.90 Using desk-top optical microscope and smart-phone-based microscope, the detection limit reached 3.86×108 exosomes/mL and 9.72×109 exosomes/mL respectively. The detection limit could be further reduced using a desk-top optical microscope.89
(3) LSPR-based biosensor
Thakur et al.91 reported an LSPR sensor for tumor-derived extracellular vesicles detection and isolation, based on self-assembly gold nanoislands (SAM-AuNIs). This work has strong potential to be extended to liquid biopsy applications in cancer diagnosis and prognosis. SAM-AuNIs is a randomly distributed nanosturcture, which is different from regular pattern nanostructures such as nano rods. Compared to the regular pattern nanostructure, the fabrication for randomly distributed nanostructures is simpler and costs less. Exosomes and microvesicles (MVs) isolated from two human cancer cell lines SH-SY5Y, A549 as well as mouse lung cancer blood serum were used as model analytes. The work demonstrated that the MVs have a weaker interaction towards SAM-AuNIs since they were removed by buffer solution flowing force within 30 mins while exosomes were stably retained with SAM-AuNIs. The sensor detection limit of exosomes was 0.194 µg/mL, and the dynamic range was 0.194–100 µg/mL. The work demonstrated exosome detection via a LSPR sensor without antibody recognition because of the specific biophysical interactions between exosomes and SAM-AuNIs.91
2) Electrochemical biosensors for exosome detection
Zhou et al.92 constructed an aptamer-based electrochemical biosensor using square wave voltammetry (SWV). In the work, the aptamer targeting CD63, an exosome transmembrane protein, was immobilized on the gold electrode surface. A probe strand with a pre-labeled redox moiety (methylene blue, MB) at one end, was hybridized with part of the anti-CD63 aptamer. When the aptamer recognizes exosomes, the probe strands with MB were stripped off from anti-CD63 aptamer, which interrupted electron transfer between MB and gold electrode surfaces and lead to a decrease in current. In the study, the biosensor showed a detection limit of 1×106 particles/mL, which was 100-fold lower than commercial immunoassays, with a linear range extending to 1×108 particles/mL.92
3) Discussion for exosome detection
The comparison of different biosensors for exosome detection is summarized in Table 4. The SERS-based biosensor88 shows sensitive detection. However, the sample preparation, exosome enrichment via magnetic beads, take more than 10 hours. The PIA-based biosensor89 utilizes a highly integrated, portable ring hole PIA biochip, which is cost-effective, real-time, and label-free. However, the detection limit is relatively high. The LSPR biosensor91 employs SAM-AuNIs to enable exosome isolation. However, the noise caused by non-specific binding is a significant challenge for clinical applications. The aptamer-based biosensor92 enables easy readout without labeling or washing and exhibits a better detection limit than conventional immunoassays for exosome detection. Some of the sensors exhibit good detection limits and wide dynamic ranges by utilizing magnetic enrichment and enzymatic amplification.
CONCLUSION
As we advance into the point-of-care era, the need for fast, specific and sensitive biosensors is growing rapidly. Biosensors for nucleic acid detection are having great potential in discovering genetic information from limited sample. More and more researchers are focusing on applying biosensors to gene tests, disease screening, and non-invasive diagnostics. Compared with nucleic acid detection, exosome detection is an emerging area which attracts attention from biosensor researchers. Exosomes, as intercellular messengers, are playing dominant roles in many pathological progresses such as metastasis. The detection and analysis of disease-related exosomes can undoubtedly facilitate their diagnosis. However, the applications of biosensors in clinical settings are facing some common challenges. Sample preparation is a significant issue. Magnetic beads have been used to pre-purify the target analyte prior to sensing,88 Although, other recently developed approaches such as temperature-responsive binary reagent system from our group93949596 have demonstrated faster and more efficient analyte enrichment than the magnetic beads sample preparation. These approaches will likely need to be integrated into the sensing process to enable rapid and specific detection and enrichment of biomarkers of interest in complex clinical samples. In addition to better sensitivity and specificity, future biosensors should also be rapid, easy-to-use, and cost effective.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download