Abstract
Objectives
The present study aimed at investigating the potential of using 18β-glycyrrhetinic acid against the cariogenic characteristics of Streptococcus mutans UA159.
Methods
The effects of 18β-glycyrrhetinic acid on biofilm formation and acid production were evaluated; the latter are indicators of cariogenicity of S. mutans. Biofilm architecture was also analyzed by scanning electron microscopy (SEM), and changes in gene expression related to biofilm formation were studied by quantitative RT-PCR.
Results
Treatment with 18β-glycyrrhetinic acid at a concentration of 20 μg/ml inhibited biofilm formation by 95% in the absence of sucrose and 60% in its presence, reduced acid production by 88.8%, and significantly suppressed the gene expression of comDE, gbpB, gtfC and vicR, which are thought to be involved in the virulence of S. mutans.
References
1. Chau NP, Pandit S, Jung JE, Jeon JG. Evaluation of Streptococcus mutans adhesion to fluoride varnishes and subsequent change in biofilm accumulation and acidogenicity. J Dent. 2014; 42(6):726–734.

2. Mitchell TJ. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat Rev Microbiol. 2003; 1(3):219–230.

3. Pecharki D, Petersen FC, Assev S, Scheie AA. Involvement of antigen I/P surface proteins in Strpetococcus mutans and Streptococcus intermedius biofilm formation. Oral Microbiol Immunol. 2005; 20(6):366–371.
4. Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011; 45(1):69–86.
5. Zhiyan H, Qian W, Yuejian H, Jingping L, Yuntao J,Rui M, et al. Use of the quorum sensing inhibitor furanone C-30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. Int J Antimicrob Agents. 2012; 40:30–35.
6. De Sousa DL, Lima RA, Zanin IC, Klein MI, Janal MN, Duarte S. Effect of twice-daily blue light treatment on matrix-rich biofilm development. PLoS One. 2015; 10(7):e0131941. DOI:10.1371/journal. pone.0131941 PMID: 26230333.

7. Matesanz-Perez P, Garcia-Gargallo M, Figuero E, Bascones-Martinz A, Sanz M, Herrera D. A systematic review on the effects of local antimicrobials as adjuncts to subgingival debridement, compared with subgingival debridement alone, in the treatment of chronic periodontitis. J Clin Perodontol. 2013; 40:227–241.
8. Asl MN,Hosseinzadeh H. Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother Res. 2008; 22:709–724.
9. Geetha RV, Roy A. In-vitro evaluation of anti bacterial activity of ethanolic root extract of Glycyrrhiza glabra on oral microbes. IJDDR. 2012; 4(4):161–165.
10. Fiore C, Eisenhut M, Ragazzi E, Zanchin G, Armanini D. A history of the therapeutic use of liquorice in Europe. J Ethnopharmacol. 2005; 99:317–324.

11. Tsukiyama RI, Katsura H, Tokuriki N, Kobayashi M. Anti-bacterial activity of licochalicone A against spore-forming bacteria. Antimi-crob Agents and Chemother. 2002; 46:1226–1230.
12. Hamid R, Rotshteyn Y, Rabadi L, Parikh R, Bullock P. Comparision of alamar blue and MTT assays for high through-put screening. Toxicol In Vitro. 2004; 18:703–710.
13. Pettit RK, Weber CA, Kean NJ, Hoffmann H, Pettit GR, Tan R, et al. Microplate alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob Agents Chemother. 2005; 49:2612–2617.
14. Ciardi JE, Rosenthal AB, Bowen WH. Rapid quantitative determination of the effect of antiplaque agents and antisera on the growth, acid production and adherence of Streptococcus mutans. J Dent Res. 1981; 60:756–762.

15. Hasan S, Danishuddin M, Adil M, Singh K, Verma PK, Khan AU. Efficacy of E. officinalis on the cariogenic properites of Streptococcus mutans: a novel and alternative approach to suppress Quorum-sensing mechanism. PLoS One. 2012; 7(7):e40319. DOI:10.1371/ Journal.pone.0040319.
16. Marsh PD. Plaque as a biofilm: pharmacological principles of drug delivery and action in the sub- and supragingival environment. Oral Dis. 2003; 9:16–22.

18. Islam B, Khan SN, Naeem A, Sharma V, Khan AU. Novel effect of plant lectins on the inhibition of Streptococcus mutans biofilm formation on saliva-coated surface. J Appl Microbiol. 2009; 106:1682–1689.
19. Matsumoto M, Minami T, Sasaki H, Sobue S, Hamada S, Ooshima T. Inhibitory effects of oolong tea extract on caries inducing properties of mutans streptococci. Caries Res. 1999; 33:441–445.
20. Ajdić D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002; 99:14434–14439.
21. Oatmen TR. Preventive dentistry techniques in the treatment of dental caries and biofilm control: A Review. Honors Proj-ects. 2011; 88.
22. Matsumi Y, Fujita K, Takashima Y,Yanagida K, Morikawa Y, Matsumoto M. Controbution of glucan-binding protein A to firm and stable biofilm formation by Streptococcus mutans. Mol Oral Micro-biol. 2015; 30:217–226.
23. Kim D, Hwang G, Liu Y, Wang Y, Singh AP, Vorsa N, et al. Cran-berry flavonoids modulate cariogenic properties of mixed-species biofilm through exopolysaccharides-matrix disruption. PLoS One. 2015; 10(12):e0145844. DOI:10.1371/journal.pone.0145844 PMID: 26713438.

24. Kuramitsu HK. Virulence properties of oral bacterial: impact of molecular biology. Curr Issues Mol Biol. 2001; 3:35–36.
25. Senadheera D, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, et al. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and fif expression, biofilm formation, and genetic competence development. J Bacteriol. 2005; 187:4064–4076.
Fig. 1.
Effect of 18b-glycyrrhetinic acid on growth of S. mutans. S. mutans incubated with present and absent of 20 μg/ml 18β-glycyrrhetinic acid in a 96well plate at 37°C.
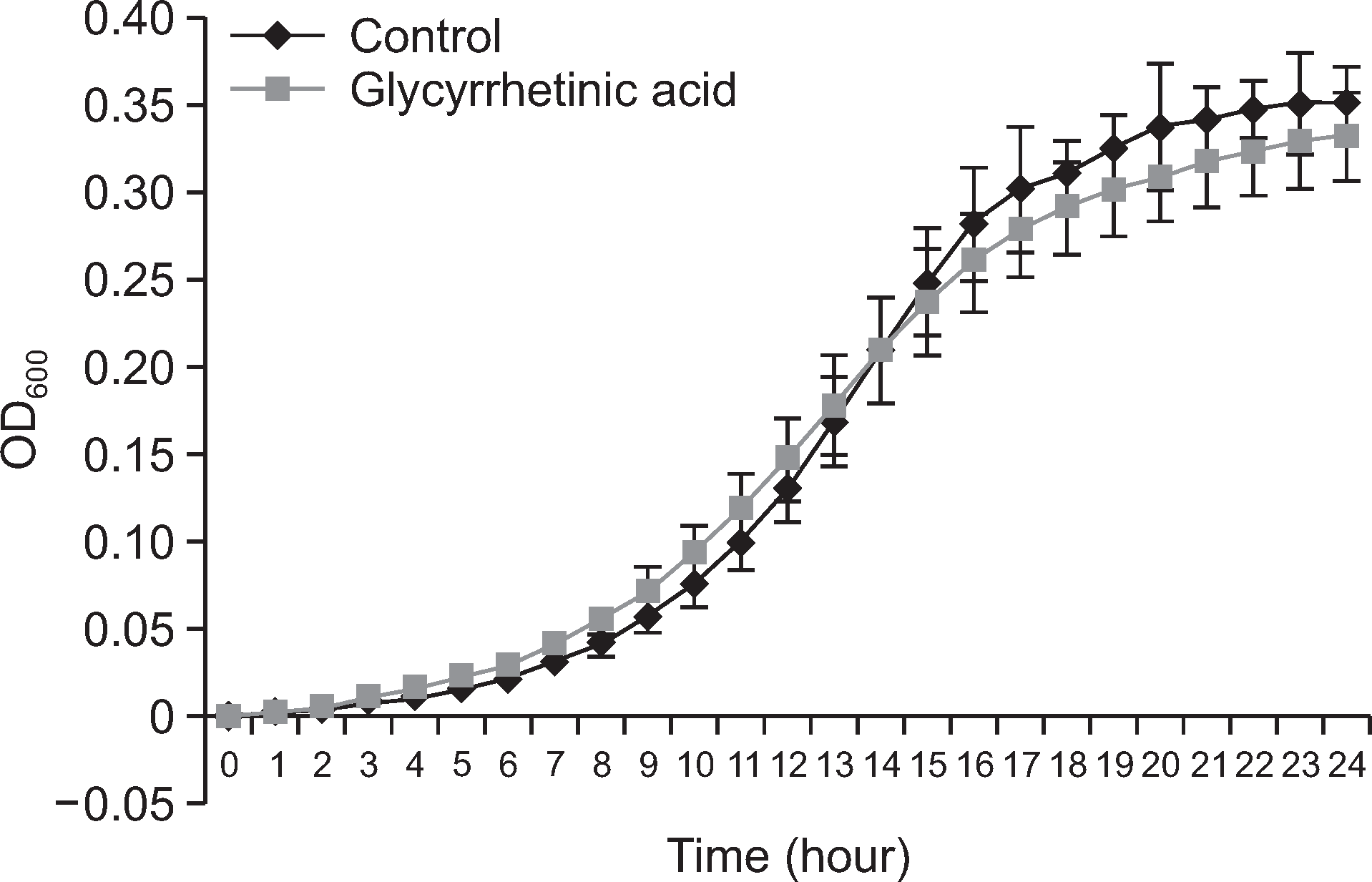
Fig. 2.
Inhibitory effect of 18β-glycyrrhetinic acid on the biofilm formation by S. mutans in the absence (sucrose-independent) and presence of 1% sucrose (sucrose-dependent). *Significant inhibition compared with untreated control (P<0.05).
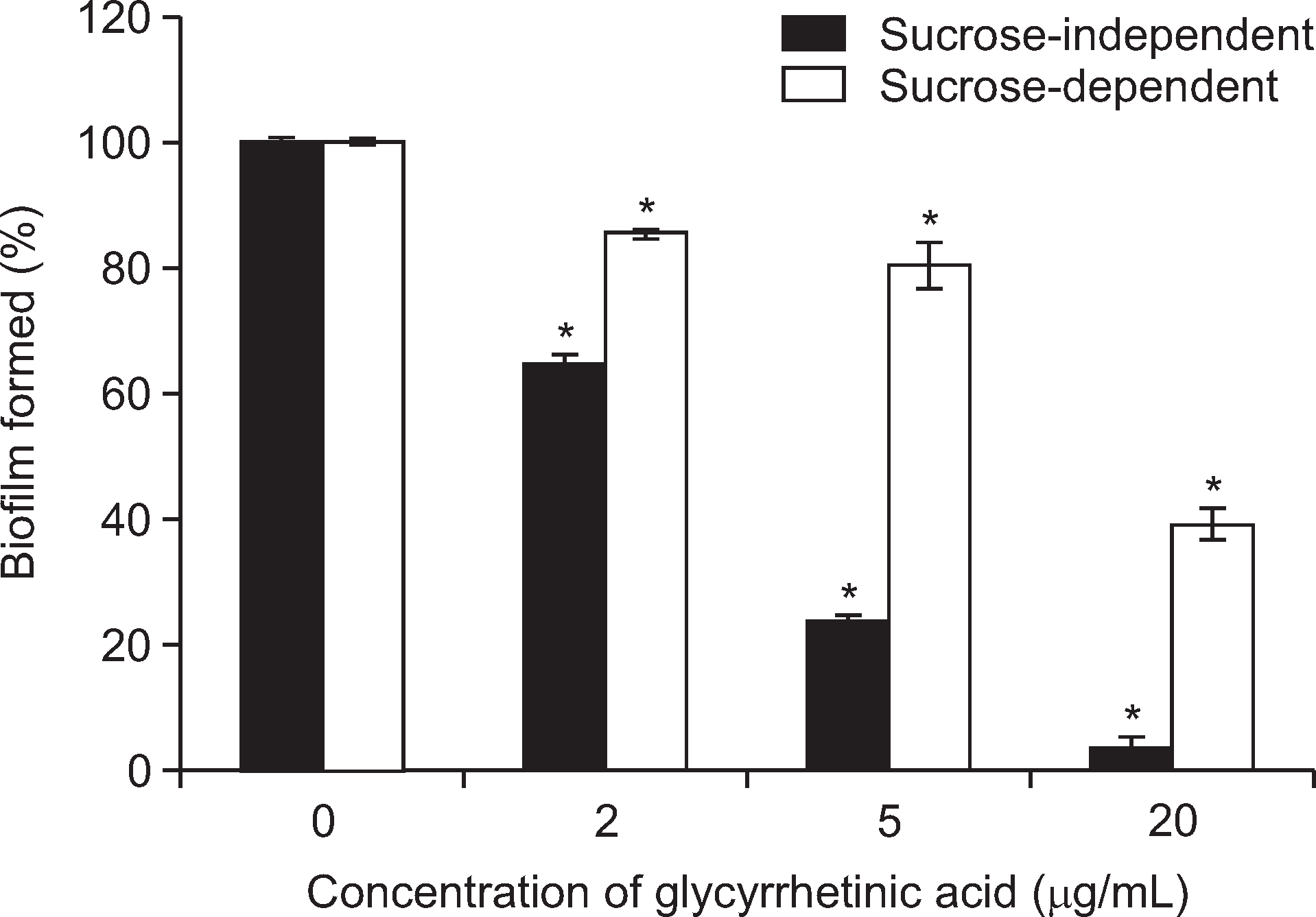
Fig. 3.
SEM images of S. mutans biofilm formed in the presence and absence of the 18β-glycyrrhetinic acid after at 37°C overnight incubation. (A) control, (B) 18β-glycyrrhetinic acid.
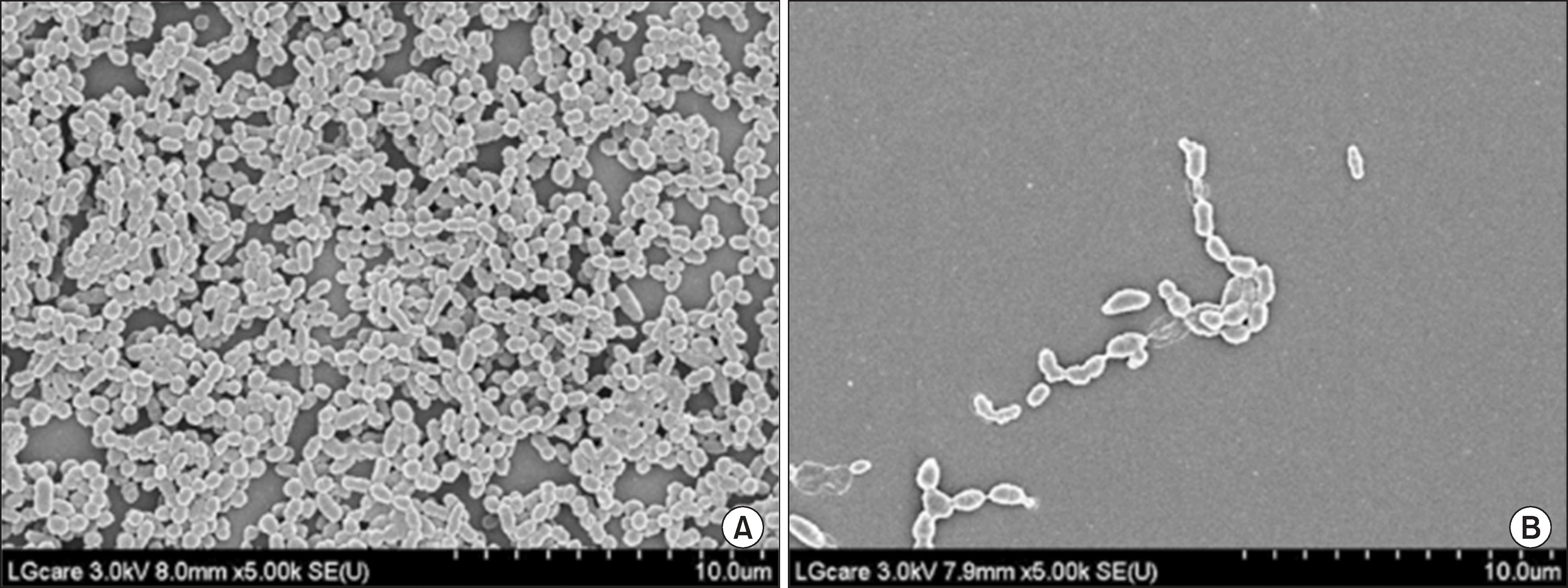
Fig. 4.
Effect of 18β-glycyrrhetinic acid on the biofilm related genes expression of S. mutans. *Significant inhibition compared with untreated control (P<0.05).
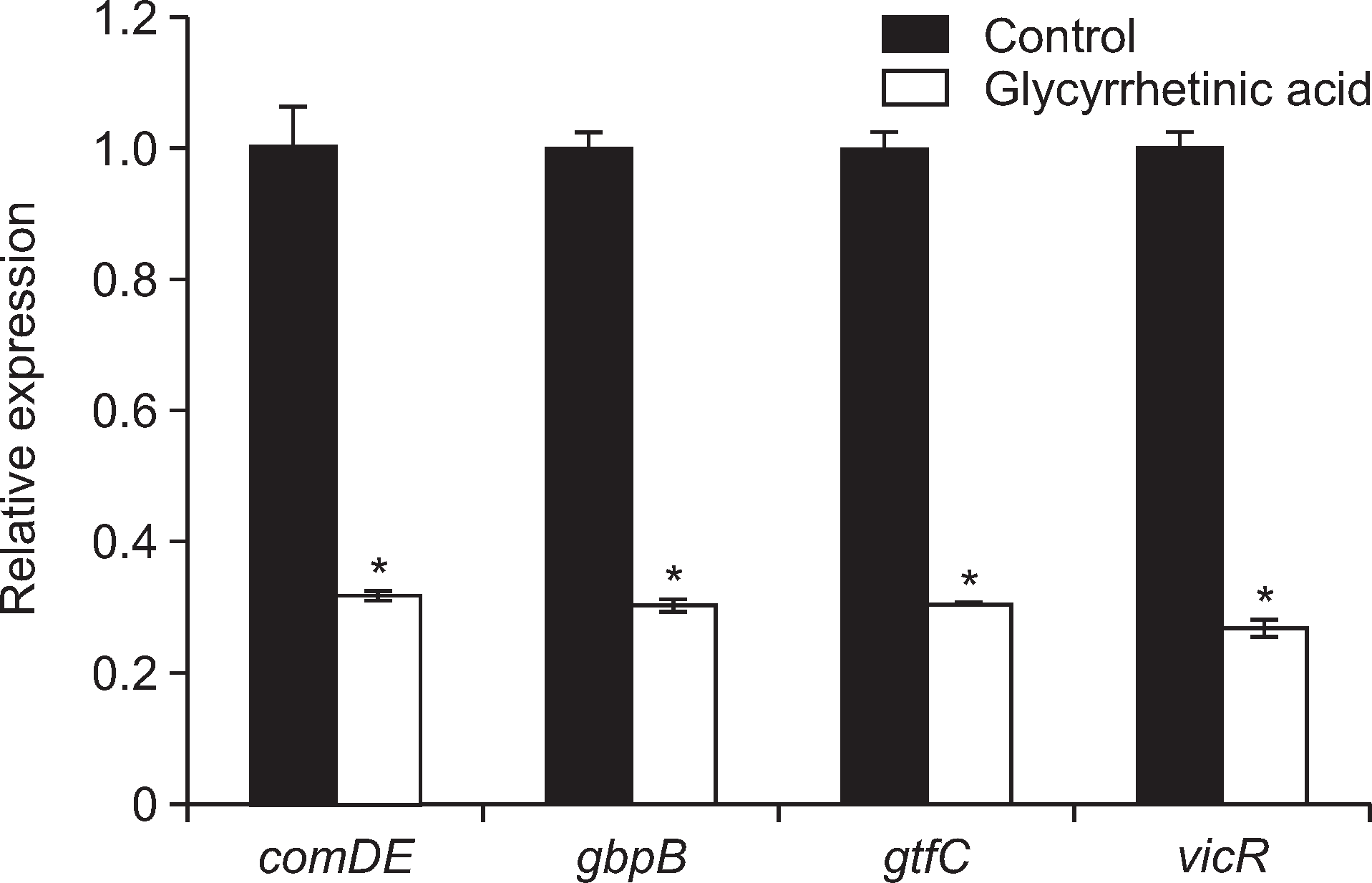
Table 1.
Nucleotide sequences of primers used in this study
| Gene† | Description |
Primer sequence (5’→3’)15) |
|
|---|---|---|---|
| Forward | Reverse | ||
| comDE | Competence-stimulating peptide | ACAATTCCTTGAGTTCCATCCAAG | TGGTCTGCTGCCTGTTGC |
| gbpB | Glucan-binding proteins (GBPs) | ATGGCGGTTATGGACACGTT | TTTGGCCACCTTGAACACCT |
| gtfC | Glucan production | CTCAACCAACCGCCACTGTT | GGTTTAACGTCAAAATTAGCTGTATTAGC |
| vicR | Two-component regulatory system | CGTCTAGTTCTGGTAACATTAAGTCCAATA | TGACACGATTACAGCCTTTGATG |
| 16S rRNA | Normalizing internal standard | CTTACCAGGTCTTGACATCCCG | ACCCAACATCTCACGACACGAG |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download