Abstract
Objective
We undertook this study to investigate the discriminant metabolites in urine from patients with established rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and from healthy individuals.
Methods
Urine samples were collected from 148 RA patients, 41 SLE patients and 104 healthy participants. The urinary metabolomic profiles were assessed using 1 H nuclear magnetic resonance spectroscopy. The relationships between discriminant metabolites and clinical variables were assessed. Collagen-induced arthritis was induced in mice to determine if a choline-rich diet reduces arthritis progression.
Results
The urinary metabolic fingerprint of patients with established RA differs from that of healthy controls and SLE patients. Markers of altered gut microbiota (trimethylamine-N-oxide, TMAO), and oxidative stress (dimethylamine) were upregulated in patients with RA. In contrast, markers of mitochondrial dysfunction (citrate and succinate) and metabolic waste products (p-cre-sol sulfate, p-CS) were downregulated in patients with RA. TMAO and dimethylamine were negatively associated with serum inflammatory markers in RA patients. In particular, patients with lower p-CS levels exhibited a more rapid radiographic progression over two years than did those with higher p-CS levels. The in vivo functional study demonstrated that mice fed with 1% choline, a source of TMAO experienced a less severe form of collagen-induced arthritis than did those fed a control diet.
Conclusion
Patients with RA showed a distinct urinary metabolomics pattern. Urinary metabolites can reflect a pattern indicative of inflammation and accelerated radiographic progression of RA. A choline-rich diet reduces experimentally-induced arthritis. This finding suggests that the interaction between diet and the intestinal microbiota contributes to the RA phenotype.
REFERENCES
3. Kuhnke A, Burmester GR, Krauss S, Buttgereit F. Bioenergetics of immune cells to assess rheumatic disease activity and efficacy of glucocorticoid treatment. Ann Rheum Dis. 2003; 62:133–9.

5. Sitton NG, Dixon JS, Bird HA, Wright V. Serum biochemistry in rheumatoid arthritis, seronegative arthropathies, osteoarthritis, SLE and normal subjects. Br J Rheumatol. 1987; 26:131–5.

6. Kapoor SR, Filer A, Fitzpatrick MA, Fisher BA, Taylor PC, Buckley CD, et al. Metabolic profiling predicts response to anti-tumor necrosis factor alpha therapy in patients with rheumatoid arthritis. Arthritis Rheum. 2013; 65:1448–56.
7. Priori R, Scrivo R, Brandt J, Valerio M, Casadei L, Valesini G, et al. Metabolomics in rheumatic diseases: the potential of an emerging methodology for improved patient diagnosis, prognosis, and treatment efficacy. Autoimmun Rev. 2013; 12:1022–30.

8. Kang MJ, Park YJ, You S, Yoo SA, Choi S, Kim DH, et al. Urinary proteome profile predictive of disease activity in rheumatoid arthritis. J Proteome Res. 2014; 13:5206–17.

9. van der Heijde D. How to read radiographs according to the Sharp/van der Heijde method. J Rheumatol. 2000; 27:261–3.
10. Bruynesteyn K, van der Heijde D, Boers M, Saudan A, Peloso P, Paulus H, et al. Determination of the minimal clinically important difference in rheumatoid arthritis joint damage of the Sharp/van der Heijde and Larsen/Scott scoring methods by clinical experts and comparison with the smallest detectable difference. Arthritis Rheum. 2002; 46:913–20.

11. Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for di-lution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006; 78:4281–90.

12. Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemom. 2002; 16:119–28.

13. Cloarec O, Dumas ME, Trygg J, Craig A, Barton RH, Lindon JC, et al. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal Chem. 2005; 77:517–26.

14. Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemom. 2006; 20:341–51.

16. Kim WU, Lee WK, Ryoo JW, Kim SH, Kim J, Youn J, et al. Suppression of collagen-induced arthritis by single administration of poly(lactic-co-glycolic acid) nanoparticles en-trapping type II collagen: a novel treatment strategy for induction of oral tolerance. Arthritis Rheum. 2002; 46:1109–20.

17. Kong JS, Yoo SA, Kim JW, Yang SP, Chae CB, Tarallo V, et al. Anti-neuropilin-1 peptide inhibition of synoviocyte survival, angiogenesis, and experimental arthritis. Arthritis Rheum. 2010; 62:179–90.

18. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011; 472:57–63.

19. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004; 25:677–86.

20. Udalova IA, Mantovani A, Feldmann M. Macrophage heterogeneity in the context of rheumatoid arthritis. Nat Rev Rheumatol. 2016; 12:472–85.

21. Seldin MM, Meng Y, Qi H, Zhu W, Wang Z, Hazen SL, et al. Trimethylamine N-Oxide promotes vascular inflammation through signaling of mitogen-activated protein kinase and nuclear factor-kappaB. J Am Heart Assoc. 2016; 5:pii: e002767.

22. Bain MA, Fornasini G, Evans AM. Trimethylamine: metabolic, pharmacokinetic and safety aspects. Curr Drug Metab. 2005; 6:227–40.

23. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015; 21:895–905.

24. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013; 2:e01202.

25. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013; 19:576–85.

26. Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018; 555:623–8.

27. Vanholder R, Bammens B, de Loor H, Glorieux G, Meijers B, Schepers E, et al. Warning: the unfortunate end of p-cresol as a uraemic toxin. Nephrol Dial Transplant. 2011; 26:1464–7.

28. Ligabue G, Damiano F, Cuoghi A, De Biasi S, Bellei E, Granito M, et al. p-Cresol and cardiovascular risk in kidney transplant recipients. Transplant Proc. 2015; 47:2121–5.

29. Yang XY, Zheng KD, Lin K, Zheng G, Zou H, Wang JM, et al. Energy metabolism disorder as a contributing factor of rheumatoid arthritis: a comparative proteomic and metabolomic study. PLoS One. 2015; 10:e0132695.

30. Biniecka M, Canavan M, McGarry T, Gao W, McCormick J, Cregan S, et al. Dysregulated bioenergetics: a key regulator of joint inflammation. Ann Rheum Dis. 2016; 75:2192–200.

31. Yang Z, Matteson EL, Goronzy JJ, Weyand CM. T-cell metabolism in autoimmune disease. Arthritis Res Ther. 2015; 17:29.

32. Tak PP, Smeets TJ, Daha MR, Kluin PM, Meijers KA, Brand R, et al. Analysis of the synovial cell infiltrate in early rheumatoid synovial tissue in relation to local disease activity. Arthritis Rheum. 1997; 40:217–25.

33. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011; 334:105–8.

34. Cho CE, Taesuwan S, Malysheva OV, Bender E, Tulchinsky NF, Yan J, et al. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition: a randomized controlled trial. Mol Nutr Food Res. 2017; 61:DOI: DOI: 10.1002/mnfr.201600324.

Supplementary Figure 1.
Variations in the levels of urinary TMAO and dimethylamine between healthy volunteers who were outliers and those who were not in the multivariate statistical analysis from Figure 1. (A), the OPLS loading plot between the two classes; (B), statistical analysis of urinary TMAO; (C), statistical analysis of urinary dimethylamine; (D), statistical analysis of urinary citrate. TMAO: trimethylamine-N-oxide, OPLS: orthogonal projections to the latent structures, a.u.: arbitrary unit.
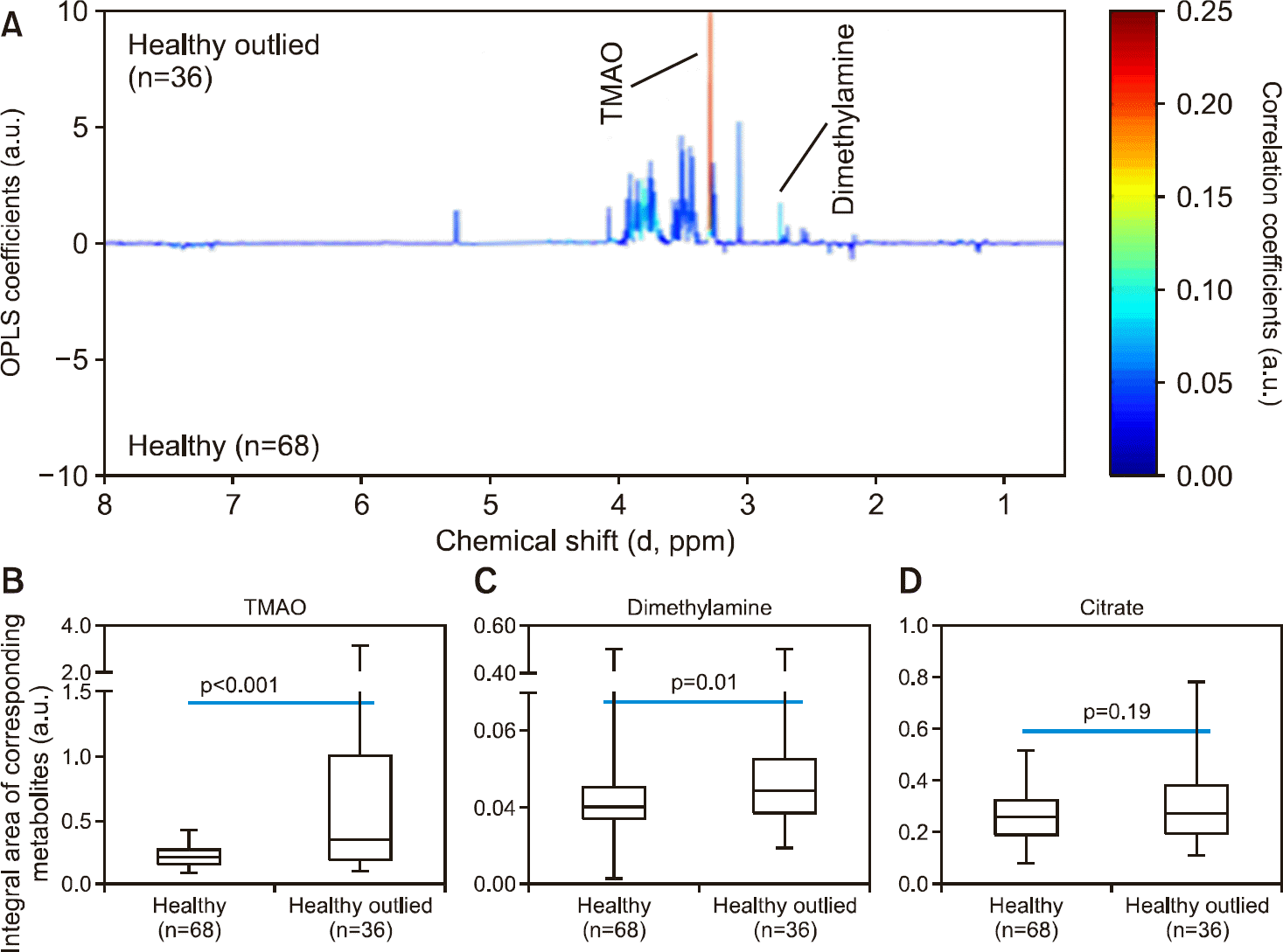
Figure 1.
Representative 1 H NMR spectrum of human urine (A). Multivariate statistical analysis including PCA (B∼ D) and OPLS-DA models (E and F, the permutation test for validation of the OPLS-DA models in the panel F) derived from 1 H NMR urinary spectra; a pairwise comparison of urinary metabolites between healthy volunteers and SLE patients (G); and between healthy volunteers and RA patients (H) in OPLS-loading plot for effective findings of urinary metabolites that differ between two classes. The OPLS-DA models in the panels (E) through (H) were displayed following removal of outliers observed in the PCA model analysis. The urinary TMAO was mainly responsible for outlying human urines including healthy volunteers and arthritis patients in multivariate statistical analysis. Panel (F) reflects validation of the OPLS-DA model in panel (E) through 200 times permutation test, in which the original Q2 value was higher than the Q2 values permuted. The upper section in the OPLS-loading plots in the panels (G and H) represent the increased urinary metabolites in SLE and RA patients compared to those of healthy volunteers. NMR: nuclear magnetic resonance, PCA: principal component analysis, OPLS-DA: orthogonal projections to the latent structures discriminant analysis, SLE: systemic lupus erythematosus, RA: rheumatoid arthritis, TMAO: trimethylamine-N-oxide, a.u.: arbitrary unit.
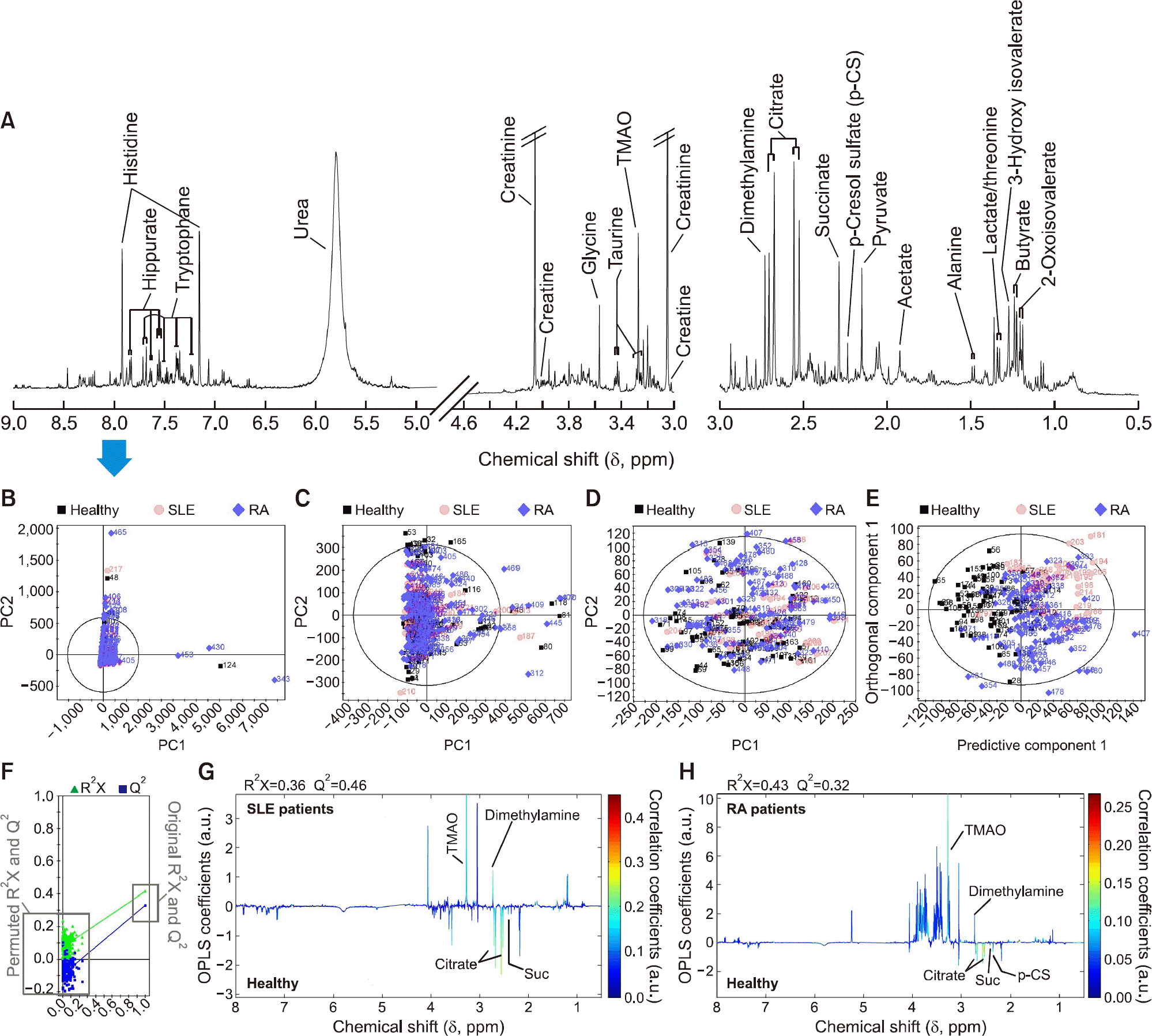
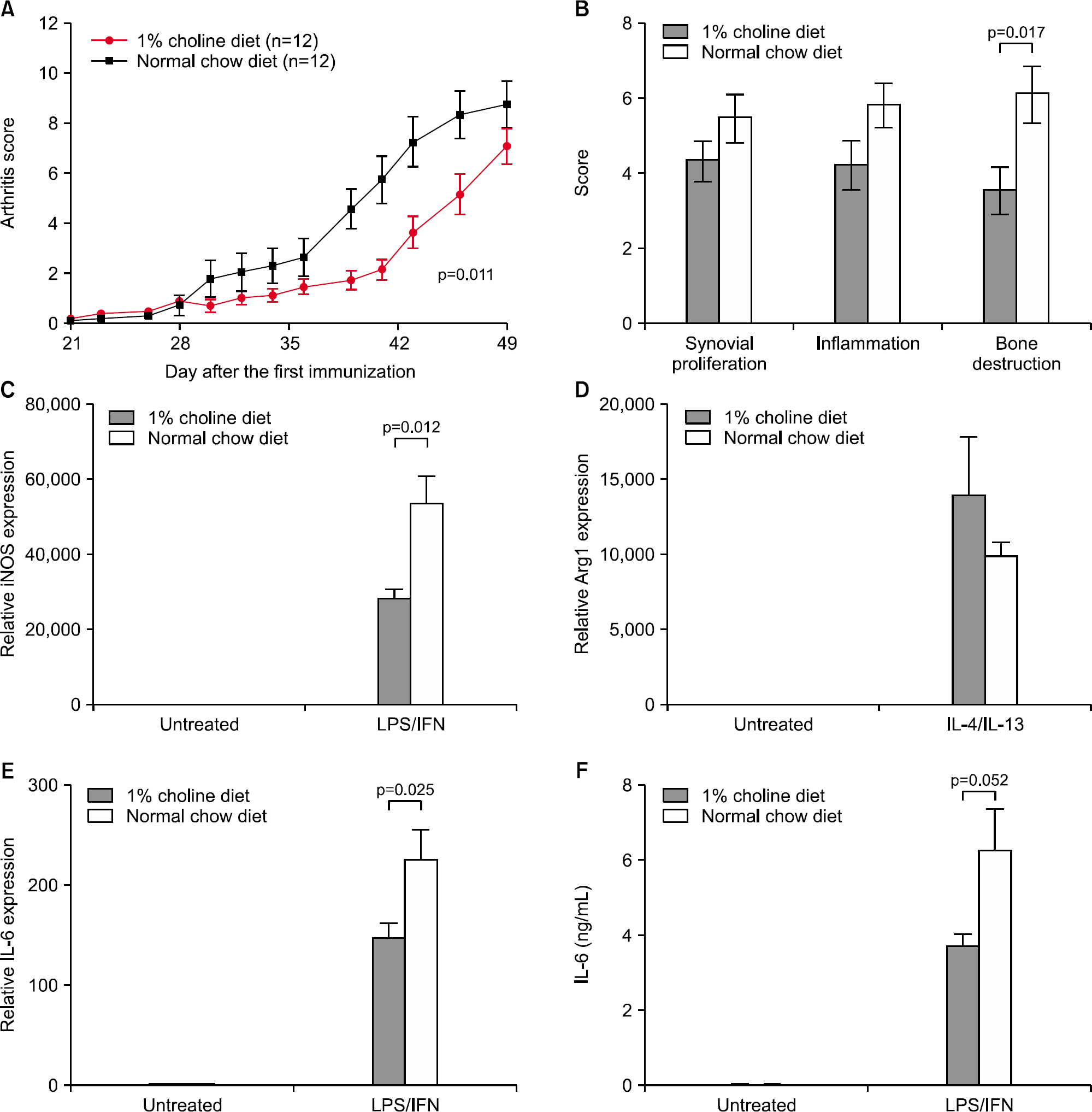
Figure 2.
Comparison of individual metabolites between healthy volunteers, patients with rheumatoid arthritis (RA), and systemic lupus erythematosus (SLE). Data in panels (A, C, E, G, and H) were displayed following removal of outliers observed in the principal component analysis (PCA) model analysis. All samples, including the outliers, removed from the PCA models were included in the statistical analysis in panels (B, D, and F). TMAO: trimethylamine-N-oxide, a.u.: arbitrary unit. *p<0.05, **p<0.01, ***p<0.001 vs. healthy volunteers.
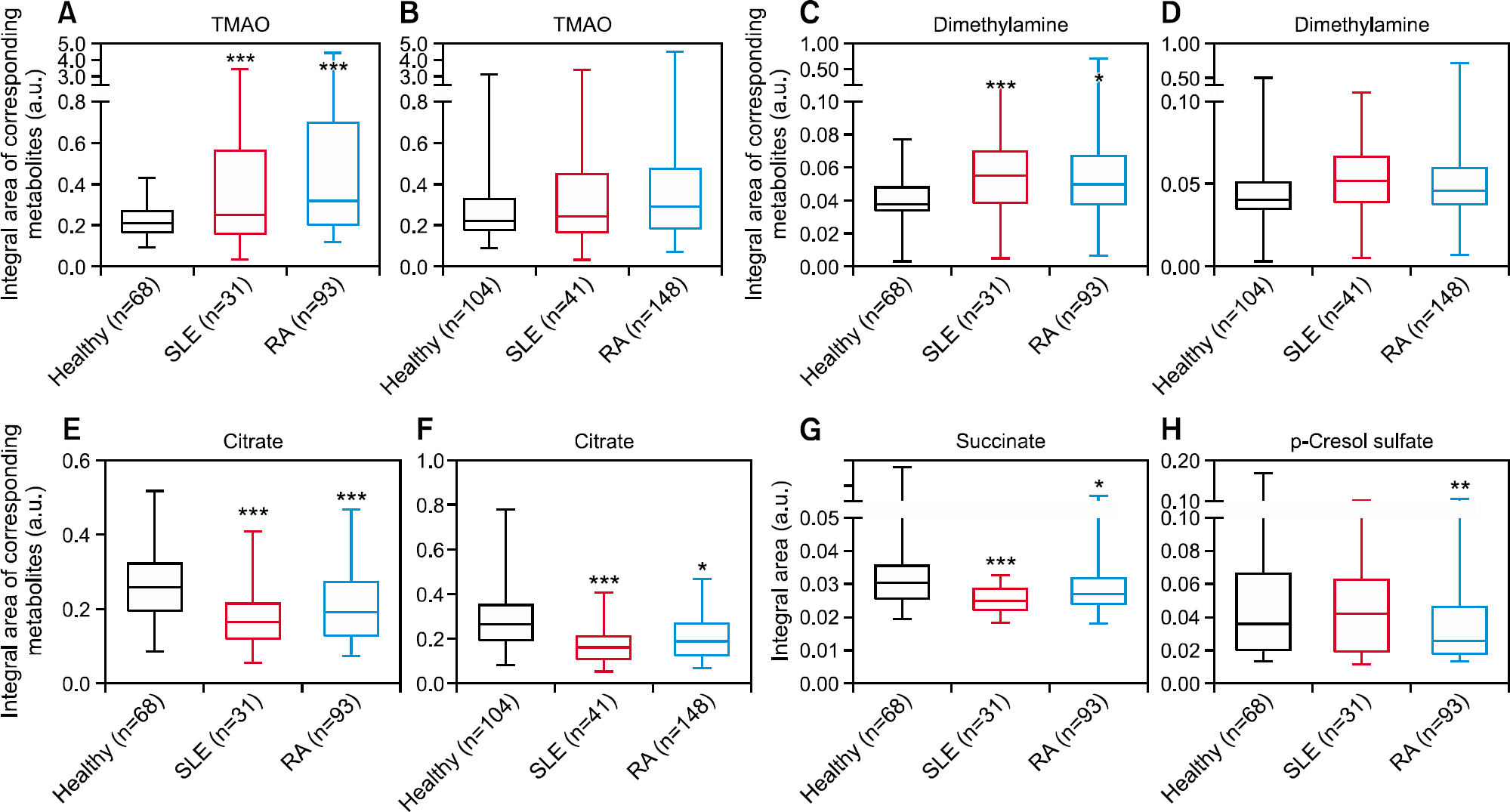
Figure 3.
Associations between urinary metabolites, inflammation and radiographic progression in rheumatoid arthritis (RA) patients.(A) Cluster correlation analysis. The power of the correlation is represented by the color and size of the circles. (B) Correlation of urinary metabolite levels with clinical inflammatory variables measured in RA patients. (C) Comparison of urinary metabolite levels in patients with radiographic progression (n=49) and without progression (n=44). (D) Multivariate logistic regression analysis for predicting radiographic progression using conventional risk factors plus urinary metabolites. ESR: erythrocyte sedimentation rate, DAS28: Disease Activity Score 28, CRP: C-reactive protein, IL: interleukin, WBC: white blood cells, DMA: dimethylamine, TMAO: trimethyl-amine-N-oxide, Hb: hemoglobin, HCQ: hydroxychloroquine, p-CS: p-cresol sulfate. *p<0.05 and **p<0.01.
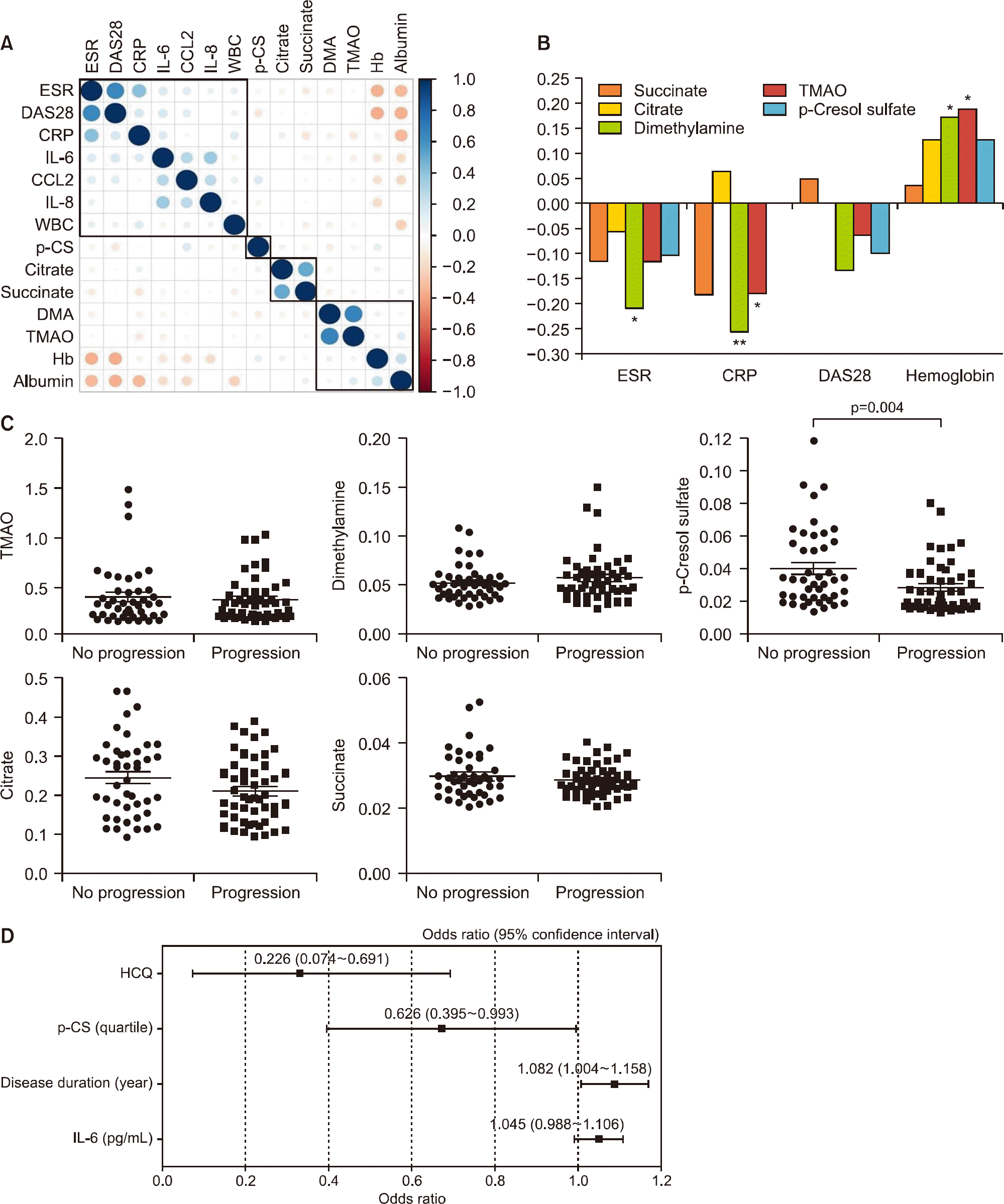
Figure 4.
Dietary choline decreases the severity of collagen-induced arthritis in mice and inhibits macrophage activation. (A) Arthritis development was assessed by measuring the arthritis score. The p-value was calculated by repeated measures ANOVA using the Greenhouse-Geisser correction. (B) Quantification of histologic mouse arthritis score. (C to F) BMDMs were stimulated with interferon (IFN)-γ (50 ng/mL) and lipopolysaccharide (LPS) (100 ng/mL) or interleukin (IL)-4 (10 ng/mL) plus IL-13 (10 ng/mL) for 24 hours. mRNA was extracted from total cell lysates and analyzed by quantitative polymerase chain reaction (qPCR) for inducible nitric oxide synthase (iNOS) (C), Arginase 1 (Arg1, D), and IL-6 (E) expression. Supernatants were analyzed by enzyme-linked immunosorbent assay (ELISA) for IL-6 (F). The data are represented by means±standard errors of the mean of 10 independent experiments.
Supplementary Table 1.
Sequences of the gene specific primers used for real-time PCR
Table 1.
Baseline patient characteristics by group
| Variable | Healthy controls (n=68) | s Patients with SLE (n=31) | Patients with RA (n=93) | p-value (HC vs. SLE) | p-value (HC vs. RA) | p-value (SLE vs. RA) |
|---|---|---|---|---|---|---|
| Age (yr) | 54 (47∼57) | 41 (34∼49) | 52 (45∼61) | <0.001 | 0.916 | <0.001 |
| Female | 65 (95.6) | 30 (96.8) | 76 (81.7) | >0.999 | 0.008 | 0.042 |
| Hypertension Diabetes mellitus | 16 (23.5) 4 (5.9) | 5 (16.1) 1 (3.2) | 24 (25.8) 6 (6.4) | 0.496 0.576 | 0.445 0.882 | 0.957 0.500 |
| Symptom duration (y | yr) NA | 4 (2∼7) | 5 (2∼10) | NA | NA | 0.354 |
| RF positive* | NA | NA | 62 (66.7) | NA | NA | NA |
| ACPA positive* | NA | NA | 74 (79.6) | NA | NA | NA |
| ESR (mm/hr) | NA | NA | 28 (19∼47) | NA | NA | NA |
| CRP (mg/dL) | NA | NA | 0.3 (0.1∼1.6) | NA | NA | NA |
| Prednisolone | NA | 22 (70.9) | 79 (84.9) | NA | NA | 0.561 |
| NSAIDs | NA | 9 (29.0) | 45 (48.4) | NA | NA | 0.027 |
| Methotrexate | NA | 5 (16.1) | 54 (58.1) | NA | NA | <0.001 |
| Hydroxychloroquine | e NA | 24 (77.4) | 64 (68.8) | NA | NA | 0.015 |
| Anti-TNF-α | NA | NA | 11 (11.8) | NA | NA | NA |
Data are presented as medians (interquartile ranges) or numbers (percentages). SLE: systemic lupus erythematosus, RA: rheumatoid arthritis, HC: healthy controls, RF: rheumatoid factor, ACPA: anti-cyclic citrullinated peptide antibody, ESR: erythrocyte sedimentation rate, CRP: C-reactive protein, NSAIDs: non-steroidal anti-inflammatory drugs, TNF-α: tumor necrosis factor-α, NA: not applicable.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download