Abstract
No study has described Streptococcus dysgalactiae subsp. equisimilis (SDSE) isolates that cause repetitive infections (recurrence and reinfection). We compared the microbiological characteristics of SDSE causing repetitive infections with those causing single infections. Three patients with invasive infections were identified based on their medical records, and multiple SDSE isolates were collected at intervals over three weeks, using a laboratory repository. Isolates from 12 patients with single-episode infections served as controls. Six isolates were collected from three patients with first and second episodes of infection. All isolates causing either repetitive or single-episode infection were subjected to emm typing, multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and random amplified polymorphic DNA (RAPD) analyses. Amplification of five virulence genes (sicG, prtF1, prtF2, lmb, and cbp), biofilm formation (BF), and cell invasion abilities (CIAs) were measured as virulent phenotypes. We observed close genetic similarities in the data obtained by emm typing, MLST, PFGE, and RAPD in four isolates from two patients, suggesting recurrence, whereas two isolates from one patient indicated genetic differences in these data, suggesting re-infection. The presence of the five virulence genes and the BF and CIA measurements appeared not to contribute to repetitive infections, compared with isolates causing single-episode infection. In conclusion, clinicians encountering patients with repetitive infections should be aware of both possibilities: recurrence with closely related strains and reinfection with different strains.
Clinical manifestations resulting from β-hemolytic streptococcal infections are well-known examples of recurring cellulitis. Nielsen et al. [1] presented a two-year survey of bacteremic episodes affected by different Lancefield groups of β-hemolytic streptococci and reported a case of recurring infection with group C streptococci. A retrospective observational study in Taiwan identified 92 patients with group G streptococcal bacteremia, with infection recurring in nine patients (incidence, 9.8%) [2]. Recently, a case of recurrent Streptococcus dysgalactiae subsp. equisimilis (SDSE) bacteremia followed by streptococcal toxic shock syndrome in a patient with Noonan syndrome was documented in Japan [3]. Trell et al. [4] performed clinical trials on patients with recurrent S. dysgalactiae bacteremia (N=22) and controls with a single episode of S. dysgalactiae bacteremia (N=92). Their case-control study showed comparable demographics, Charlson comorbidity scores, and clinical presentations. Importantly, no study has described the microbiological characteristics of SDSE isolates causing repetitive infections (including recurrence and reinfection) during different clinical courses of the same patients in comparison with those having single episodes during the observation period. Therefore, we compared the phenotypic and genotypic characteristics of SDSE strains causing repetitive infections with those causing single infections. Our findings would be useful for clinicians and microbiologists.
We retrospectively retrieved medical information of 15 patients (median age=82 years, range=64 to 88 years, 6 males and 9 females) with invasive SDSE infection because such patients possess the possibility for repetitive onsets [1234]. The information pertained to underlying medical conditions, clinical diagnoses, laboratory test data while obtaining bacterial cultures, therapeutic antimicrobial agents, surgical interventions, and outcomes. The presence of invasive SDSE infection was determined using cultures from sterile sites (multiple sets of blood cultures) [5]. Repetitive infections were divided into recurrence (caused by isolates with molecular epidemiological findings similar to the previous episode) and reinfection (caused by isolates with different molecular epidemiological findings from the previous infection). Death from the infection within three weeks of disease onset or disease-associated sequelae following the infection was considered a poor outcome [5]. Additionally, we retrospectively confirmed patients with single-episode invasive infections during the same time period and used their isolates as controls. Written informed consent was obtained from the patients at admission. Our study protocol was approved by the ethics committee of Kitasato University Medical Center, Saitama, Japan (Approval No. 29-6).
We collected six isolates with β-hemolytic groups G/C/A streptococcal infection at intervals over three weeks from a repository at the Clinical Laboratory of Kitasato University Medical Center from May 1, 2014 through April 30, 2017 as these isolates might cause repetitive infections. These isolates were identified as SDSE using an API-20 Strep system (Sysmex BioMérieux, Tokyo, Japan) for biochemical testing, followed by confirmation using PCR amplification of the 16S rRNA gene, as described previously [6]. The isolates were considered positive if the PCR-amplified product yielded at least one sequence showing ≥98.7% similarity with the 16S rRNA sequence of the type strain National Collection of Food Bacteria (NCFB) 1356(T). All SDSE isolates were stored at −70℃ to −80℃ until further evaluation. Additionally, we included American Type Culture Collection (ATCC) 12394 (G group strain D166B) as a quality control.
All isolates were subjected to emm genotyping, multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and random amplified polymorphic DNA (RAPD) analyses, as described previously [67]. Briefly, all emm typing was based on the U.S. Centers for Disease Control and Prevention database (http://www2a.cdc.gov/ncidod/biotech/strepblast.asp). MLST was performed by sequencing seven housekeeping genes (gki, gtr, murI, mutS, recP, xpt, and atoB) according to the SDSE website (http://pubmlst.org/sdysgalactiae/). Sequence types (STs) were grouped into clonal complexes, in which related STs were classified as single locus variants differing in only one housekeeping gene [678]. PFGE profiling following DNA digestion with the restriction enzyme SmaI was also performed [6]. RAPD analysis was carried out using three different primers (H2, P5, and P6) [7]. The streptococcal inhibitor of complement-mediated cell lysis-like gene (sicG) was amplified, and the amplicons were sequenced [67]; four genes (prtF1, prtF2, lmb, and cbp) encoding adhesin factors were also amplified [9].
Furthermore, the biofilm formation (BF) of all isolates causing repetitive/single-episode infection was measured with minor modifications (absorbance at 545 nm and comparable bacterial loads [1×105 colony-forming units (CFU)/well] based on growth curves when starting the cultures) as described previously [9]. To examine additional virulent factors, we measured the cell invasion abilities (CIAs) as described previously with slight modifications (multiplied infection of 10 SDSE per one host cell, no centrifugation was performed, and non-centrifuged SDSE isolates were added onto semi-confluent human colon carcinoma cells [Caco-2] grown in a 12-well culture plate with minimum essential medium supplemented with 15% fetal calf serum) [10]. Resistance to antimicrobials was determined using the broth microdilution method according to the CLSI guidelines for β-hemolytic streptococci [11]. The presence of antimicrobial resistance genes, including erm(A), erm(B), mef(A), tet(M), tet(O), tet(K), tet(L), and tet(S), was confirmed in the resistant isolates [678].
BF and CIA data were summarized as mean (fold value/mean of ATCC strain)±SD of five wells and mean (fold value/mean of ATCC strain)±SD of four wells, respectively. Data were compared using Welch's t-test following the F-test. Statcel4 (OMS Publisher, Tokyo, Japan) was used for all analyses. P<0.05 indicated statistical significance.
Based on the charts, of the 15 patients, three developed repetitive infections without another infection due to pathogens other than SDSE (incidence, 20%). A total of six isolates were extracted from the repository (Table 1). None of the three patients showed poor outcomes or sequential repetitive infections as of July 31, 2018. The characteristics of the six isolates causing repetitive infections and strain ATCC 12394 are shown in Table 1. The other 12 patients showed single-episode infection, and the characteristics of the 12 isolates are provided in Supplemental Data Table S1.
emm typing, MLST, PFGE, and RAPD analyses using the DNA samples from KM32/KM32-2 and KM36/KM36-2 revealed close genetic similarities (Table 1 and Fig. 1), suggesting recurrent strains of KM32-2 and KM36-2. Analyses using the DNA samples from KM10/KM10-2 showed different genotypic characteristics, suggesting reinfection by a KM10-2 strain. The DNA-profile of five virulence genes (sicG, prtF1, prtF2, lmb, and cbp), as well as the BF and CIA phenotypes of the isolates causing repetitive infections, did not appear to significantly contribute to the recurrence/reinfection mechanisms, as compared with the isolates causing single-episode infection.
Lo et al. [12] reported the prevalence of several virulence genes in 246 SDSE isolates from central Taiwan during 2007–2011. The distributions of cbp, fbp (encoding fibronectin-binding protein), and sicG were 14.6%, 9.4%, and 2.4%, respectively, whereas that of lmb was 100%. Another study suggested that the presence of genes encoding fibronectin-binding protein (prtF1, prtF2) constitutes a potential virulence factor necessary for the SDSE strains to colonize the human body [9]. Therefore, we evaluated the presence of sicG, prtF1, prtF2, and cbp in the repetitive infection isolates, while amplification of lmb was included as a positive control. However, the presence of these genes, as well as the BF and CIA phenotypes, did not appear to serve as recurrent factors. Thus, there were limitations regarding the evidence of factors (DNA profile of adhesin factors as well as sicG and BF/CIA) that significantly contributed to the repetitive infections in our study.
We did not determine the colonization site, unlike a previous study that identified anal/perianal streptococcal colonization in cases of erysipelas and cellulitis, using samples from the anal canal of patients with repetitive infections [13]. In our study too, the colonization site may be the anal canal, because our patients were mostly elderly (median age: 75 years), and such individuals have poor anal function [5].
In summary, we identified two patients with SDSE recurrence and one patient with reinfection based on data from emm typing, MLST, PFGE, and RAPD analyses. In agreement with our observations, SDSE recurrence has been documented using emm typing and PFGE analysis in recurrent cases [2]. Thus, clinicians and infection control staff encountering patients developing repetitive infections should be aware of both possibilities: recurrence due to closely related strains and reinfection by different strains. Our findings regarding reinfection add to existing knowledge of SDSE infections.
Acknowledgements
This study made use of the Streptococcus dysgalactiae MLST website (http://pubmlst.org/sdysgalactiae/) of the University of Oxford (Jolley & Maiden 2010, BMC Bioinformatics, 11:595). The development of this site has been funded by the Wellcome Trust. The authors wish to thank Editage (http://www.editage.jp) for English language editing.
References
1. Nielsen SV, Kolmos HJ. Bacteraemia due to different groups of beta-haemolytic streptococci: a two-year survey and presentation of a case of recurring infection due to eStreptococcus “quisimilis”. Infection. 1993; 21:358–361. PMID: 8132363.
2. Liao CH, Liu LC, Huang YT, Teng LJ, Hsueh PR. Bacteremia caused by group G Streptococci, Taiwan. Emerg Infect Dis. 2008; 14:837–840. PMID: 18439377.
3. Suzuki K, Nakamura A, Ishikura K, Imai H. Recurrent SDSE bacteraemia resulting in streptococcal toxic shock syndrome in a patient with Noonan syndrome. BMJ Case Rep. 2016; 2016:pii: bcr2016216092.
4. Trell K, Sendi P, Rasmussen M. Recurrent bacteremia with Streptococcus dysgalactiae: a case-control study. Diagn Microbiol Infect Dis. 2016; 85:121–124. PMID: 26906192.
5. Takahashi T, Sunaoshi K, Sunakawa K, Fujishima S, Watanabe H, Ubukata K, et al. Clinical aspects of invasive infections with Streptococcus dysgalactiae ssp. equisimilis in Japan: differences with respect to Streptococcus pyogenes and Streptococcus agalactiae infections. Clin Microbiol Infect. 2010; 16:1097–1103. PMID: 19732082.
6. Fujita T, Horiuchi A, Ogawa M, Yoshida H, Hirose Y, Nagano N, et al. Genetic diversity in Streptococcus dysgalactiae subsp. equisimilis isolates from patients with invasive and noninvasive infections in a Japanese university hospital (2014-2015). Jpn J Infect Dis. 2017; 70:100–104. PMID: 27000456.
7. Takahashi T, Fujita T, Shibayama A, Tsuyuki Y, Yoshida H. Prevalence of complement-mediated cell lysis-like gene (sicG) in Streptococcus dysgalactiae subsp. equisimilis isolates from Japan (2014–2016). Ann Lab Med. 2017; 37:297–304. PMID: 28445008.
8. Kim S, Byun JH, Park H, Lee J, Lee HS, Yoshida H, et al. Molecular epidemiological features and antibiotic susceptibility patterns of Streptococcus dysgalactiae subsp. equisimilis isolates from Korea and Japan. Ann Lab Med. 2018; 38:212–219. PMID: 29401555.
9. Ciszewski M, Szewczyk EM. Potential factors enabling human body colonization by animal Streptococcus dysgalactiae subsp. equisimilis strains. Curr Microbiol. 2017; 74:650–654. PMID: 28314902.
10. Kawabata S, Kuwata H, Nakagawa I, Morimatsu S, Sano K, Hamada S. Capsular hyaluronic acid of Group A Streptococci hampers their invasion into human pharyngeal epithelial cells. Microb Pathog. 1999; 27:71–80. PMID: 10458918.
11. CLSI. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute;2012.
12. Lo HH, Cheng WS. Distribution of virulence factors and association with emm polymorphism or isolation site among beta-hemolytic group G Streptococcus dysgalactiae subspecies equisimilis. APMIS. 2015; 123:45–52. PMID: 25244428.
13. Eriksson BK. Anal colonization of group G beta-hemolytic streptococci in relapsing erysipelas of the lower extremity. Clin Infect Dis. 1999; 29:1319–1320. PMID: 10524984.
SUPPLEMENTARY MATERIAL
Supplemental Data Table S1
Characteristics of SDSE isolates causing invasive infection with a single episode
Fig. 1
Analyses of SDSE isolates causing repetitive infections: (A) PFGE and (B) RAPD DNA. We used restriction enzyme SmaI and three different (H2, P5, and P6) primers.
Abbreviations: PFGE, pulsed-field gel electrophoresis; RAPD, random amplified polymorphic DNA; SDSE, Streptococcus dysgalactiae subsp. Equisimilis; M, marker; A, ATCC 12394; 1, KM10; 2, KM10-2; 3, KM32; 4, KM32-2; 5, KM36; 6, KM36-2.
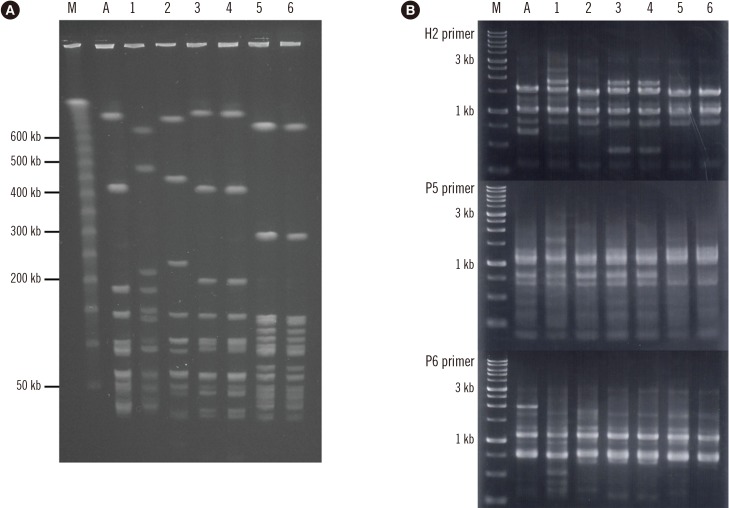
Table 1
Characteristics of SDSE isolates causing repetitive infections (recurrence and reinfection)
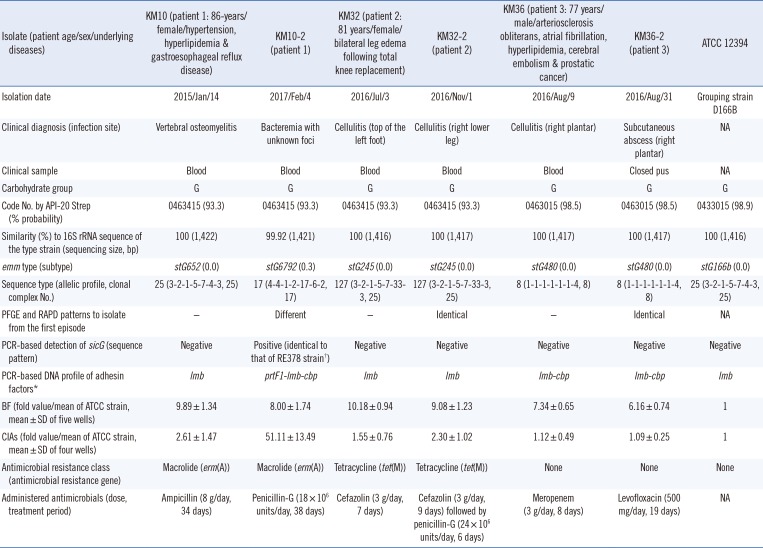
*The adhesin factors include fibronectin-binding protein 1/2, laminin-binding protein, and collagen-binding protein; †Strain RE378 (SDSE isolates in Japan) contains the sequence of the streptococcal inhibitor of complement-mediated cell lysis-like gene.
Abbreviations: SDSE, Streptococcus dysgalactiae subsp. equisimilis; PFGE, pulsed-field gel electrophoresis; RAPD, random amplified polymorphic DNA; sicG, streptococcal inhibitor of complement-mediated cell lysis-like gene; BF, Biofilm formations; CIA, cell invasion abilities; SD, standard deviation; NA, not available.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download