Abstract
Background
Negative urine cultures to rule out urinary tract infections (UTI) generate a considerable laboratory workload; thus, a rapid screening test is desirable. We evaluated the performance of a new automated microscopy analyzer, cobas u 701 (Roche Diagnostics International, Rotkreuz, Switzerland) for the screening of UTI, and developed a rule-out strategy to reduce the number of samples requiring culture. We also assessed squamous epithelial cell (SEC) count as a predictor of culture contamination.
Methods
In total, 1,604 urine samples from outpatients were analyzed with cobas u 701 and culture. Bacterial (BAC) and white blood cell (WBC) counts were used for sample interpretation. To determine a useful cut-off point to predict negative cultures, we selected the highest sensitivity and specificity values obtained from ROC curves. Diagnostic accuracy by age and gender was evaluated.
Results
Urine culture showed growth of ≥104 colony forming units (CFU)/mL in 256 samples (16.0%). The highest sensitivity (91.8%) and specificity (68.4%) were obtained for cut-off points of 119 BAC/µL and 22 WBC/µL. The combination of BAC and WBC improved the performance of the rule-out strategy with a low rate of false-negative results (1.5%) and a high negative predictive value (NPV, 97.3%). Fifty-seven percent of the samples would not have required culture. SEC count was a poor predictor of culture contamination.
Urinary tract infections (UTI) are an important cause of morbidity and remain the second most common indication for antibiotic therapy in primary and secondary care [1]. The majority of UTI are treated in the ambulatory setting; thus, urine samples are among the most frequently received outpatient samples in clinical microbiology laboratories.
Quantitative urine culture, though time-consuming and laborious, is still considered the gold standard for UTI diagnosis, but as many as 70–80% of requested urine cultures, mainly in the ambulatory care setting, prove to be negative [23], Negative urine cultures generate a considerable workload and not only cause a waste of resources but also delay other tests pending in a clinical laboratory. Thus, a rapid screening test to rule out UTI is desirable.
The use of flow cytometry-based analyzers that quantitatively measure both bacterial (BAC) and white blood cell (WBC) counts has been evaluated for screening purposes, with variable results [24567]. Urine sediment analysis by manual microscopy is an efficient screening method but suffers from interobserver variability and is labor-intensive [8]. Automated microscopy is extensively used for urine sediment analysis and has recently been evaluated for bacteriological screening of urine samples, with high accuracy in terms of sensitivity and negative predictive value (NPV) [3910]. Additionally, as compared with urine culture, it shortens the turnaround time of analysis, and may reduce both workload and laboratory costs [9].
cobas u 701 (Roche Diagnostics International, Rotkreuz, Switzerland) is a recently introduced automated urine sediment analysis system. It provides semiquantitative analysis of BAC; quantitative analysis of WBC count, squamous epithelial cell (SEC) count, non-SEC, and hyaline casts; and qualitative analysis of pathological casts, crystals, yeast, sperm, and mucus [11]. To date, only Kim et al. [12] have assessed its usefulness in the screening of UTI, reporting promising results. Although the reported overall diagnostic accuracy was good, this study did not provide either the level of care from which the urine samples were obtained or specific results on test performance according to age and gender. To determine the real performance of this analyzer for the screening of UTI, large studies that include samples from patients with different characteristics are needed. As cobas u 701 provides semiquantitative analysis of SEC, and their presence has been related to urinary contamination, it is important to determine the value of SEC count as a predictor of urine culture contamination. This was not addressed by Kim et al. [12].
We evaluated the analytical performance of cobas u 701 in the microbiological screening of urine samples from a large population of outpatients. We also developed a screening strategy to reduce the number of urine samples requiring culture, while maintaining a low rate of false negatives and a high NPV. Additionally, we assessed SEC count as a predictor of culture contamination because there is limited evidence supporting this association.
This prospective study was carried out at the Microbiology Service of San Juan University Hospital, Alicante, Spain. For this type of study, no ethics committee approval or informed consent was required in our institution. Urine samples were randomly selected from all samples submitted for culture to the general microbiology laboratory to rule out UTI from outpatient community collection sites between September and November 2017. Samples from children and pregnant women were excluded. Forty-two samples were discarded because of visible blood or high turbidity. In total, 1,604 samples were finally included: 1,322 (82.4%) from women (mean age: 49.3±21.5 years) and 282 (17.6%) from men (mean age: 61.5±18.3 years). The distribution by age group was as follows: 14–19 years, N=57; 20–49 years, N=642; ≥50 years, N=661. Samples were collected in preservative tubes (Vacutainer Plus C&S Boric Acid; Becton Dickinson Diagnostics Systems, Franklin Lakes, NJ, USA) and examined within six hours of receipt.
Samples were inoculated using a 1-µL loop on Columbia blood agar and MacConkey agar plates. Quantitative growth was assessed after 18–24 hours of incubation at 37℃. A BAC count ≥104 colony forming units (CFU)/mL was considered positive, and no growth or <104 CFU/mL was considered negative. Three or more isolates without a dominant pathogen was considered to represent contamination. Bacterial identification was carried out using an automated system (WalkAway system, Beckman Coulter, West Sacramento, CA, USA) or conventional biochemical methods.
The sample tubes were placed in racks and introduced into the analyzer, where 200 µL of urine was transferred to a cuvette and centrifuged at 260×g for 10 seconds. A built-in camera takes 15 digital images from different locations within the cuvette. All particles in the 15 digital images represent those contained in 2.3 µL of native urine. The images are evaluated using the high-quality image processing software Auto Image Evaluation Module (AIEM) (version 8, Roche Diagnostics International). cobas u 701 can analyze 40 samples in 35 minutes. We used BAC, WBC, and SEC counts.
The diagnostic accuracy of cobas u 701 was evaluated for the entire population and for subgroups based on patient age and gender. Positive predictive value (PPV) and NPV of BAC and WBC counts were compared against the urine culture results. For testing, samples were divided into two groups: negative and positive by culture. Diagnostic accuracy of BAC and WBC counts was measured using the area under the ROC curve (AUC) with a two-sided 95% confidence interval (CI). To determine a useful cutoff point to predict negative cultures, we selected the highest sensitivity and specificity values obtained from the ROC curves. We also evaluated whether BAC and WBC counts combined would increase the sensitivity and NPV. In addition, an ROC curve was generated to evaluate the predictive value of SEC count for urine sample contamination. We analyzed the data using R 3.4.3 (R Development Core Team, 2017).
Culture showed no growth or BAC <104 CFU/mL in 1,104 (68.8%) samples (900 from women and 204 from men). BAC ≥104 CFU/mL was observed in 256 (16.0%) samples (221 from women and 35 from men). In total, 244 (15.2%) cultured samples were considered contaminated and were excluded from the analyses, leaving a total of 1,360 valid samples.
The microorganisms identified were typical for UTI; the most frequent causative agents were Escherichia coli (60.5%), Klebsiella pneumoniae (14.8%), Enterococcus faecalis (5.5%), Proteus mirabilis (3.1%), Streptococcus agalactiae (3.1%), Citrobacter koseri (2.0%), and Staphylococcus saprophyticus (1.2%).
Using ROC curves, we compared the BAC and WBC counts against culture results. The AUC (95% CI) for BAC count was 0.877 (0.851–0.902), and for WBC count, 0.857 (0.830–0.885). The best sensitivity and specificity values were obtained for cut-off points of 119 BAC/µL and 22 WBC/µL. The combination of BAC and WBC counts improved the accuracy of the rule-out strategy, resulting in a low rate of false-negative results and a high NPV (97.3%) (Table 1). Using this algorithm, 776 samples, accounting for 57% of the total samples, would have not required culture, although only 755 of the 776 samples would have been correctly screened (true negative results); the remaining 21 (1.5%) would have been incorrectly screened (false-negative results). The false-negative results were as follows: E. coli (5), K. pneumoniae (3), S. agalactiae (3), P. mirabilis (2), Enterococcus faecalis (2), Citrobacter koseri (1), Kluyvera ascorbate (1), Enterobacter cloacae (1), Enterobacter aerogenes (1), Candida spp. (1), and Corynebacterium urealyticum (1). Using the same algorithm, we found that 25.4% of the samples would have been cultured unnecessarily (349 false-positive results) (Table 1). The diagnostic accuracy of cobas u 701 was high in all age groups, although the performance was poorer for patients aged 14–19 years than for older patients (Table 2).
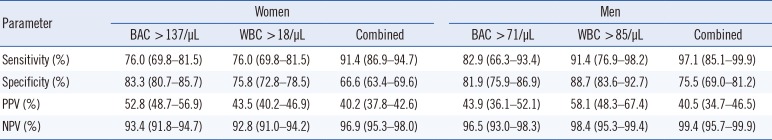
To study the diagnostic performance of both BAC and WBC counts according to gender, we generated ROC curves for women and men separately. The highest sensitivity and specificity values suggested cut-off points of >137 BAC/µL and >18 WBC/µL for women and >71 BAC/µL and >85 WBC/µL for men. By using the combination of BAC and WBC cut-off values, sensitivity and NPV improved in both women and men (Table 3). Nineteen (1.7%) false-negative samples were from women, and only one (0.4%) was from a man. Further, 55% (N=618) and 64.8% (N=155) of samples from women and men, respectively, would have not required culture.
SEC count was a poor predictor of urine sample contamination (AUC, 0.551; 95% CI, 0.512–0.591).
Our study showed that screening for UTI with cobas u 701 can reduce the number of urine samples requiring culture, with a low rate of false negatives. We developed an algorithm combining BAC and WBC counts that achieved an NPV of 97% and thus may be a very efficient strategy to rule out UTI in outpatient urine samples.
Several analyzers have been used to screen for UTI, most of them based on flow cytometry, with dissenting results. Evans et al. [5] using the UF-100 (Sysmex Corporation, Kobe, Japan) reported an NPV of 96% and a sensitivity of 92%, whereas Zaman et al. [6] concluded that the UF-100 was not suitable for UTI screening. However, comparing studies is difficult as reported sensitivities and specificities depend on the definition used for gold standard-positive and -negative urine samples. In addition, Broeren et al. [13] found that the applicability of the UF-1000i (Sysmex Corporation) to screen for negative urine samples strongly depends on the prevalence of UTI and the definition of positive and negative urine cultures used in the laboratory. Furthermore, there is an ongoing debate about the cut-off values for the best discrimination between positive and negative samples [714].
Patient characteristics may be important when assessing the performance of the automated urine analyzers. A recent study in outpatients found that the UF-1000i system was useful for applying age-specific cutoffs to screen for UTI [15]. Automated microscopy is extensively used for urine sediment analysis but has been less assessed than flow cytometry for bacteriological screening of urine samples. Recently, Falbo et al. [9] reported a sensitivity of 98% and NPV of 99% using sediMAX (Menarini Diagnostics, Florence, Italy), an automated microscopy urine analyzer, with a 47% reduction in the need for urine culture, for a population with a UTI prevalence of 18%, which is similar to that found in our study.
To our knowledge, only one previous study evaluated cobas u 701 [10]. Using a BAC count of ≥105 CFU/mL as the gold standard, the authors reported a sensitivity of 81.5% and NPV of 96.2%. On the basis of a cut-off value of ≥104 CFU/mL (the threshold considered in our study), the sensitivity and specificity were 79% and 73%, respectively. Unfortunately, the authors compared BAC and yeast counts, but they did not integrate WBC count. In our study, the best performance of the rule-out strategy (low false-negative rate and high NPV) was achieved when both BAC and WBC counts were used, reaching a sensitivity and NPV of 91.8% and 97.3%, respectively.
We believe that for bacteriological screening of urine samples, it is important for each laboratory to establish cut-off points for BAC and WBC counts, considering their UTI prevalence and the characteristics of the patients as recommended [13161718].
In the whole population, we established >119 BAC/µL and >22 WBC/µL as optimal cut-off points. With both values combined, we obtained a higher NPV for all subgroups studied, as has been also described in previous studies using sediMAX to rule out negative urine samples. For example, Martìnez et al. [10] reported an NPV of 99%; Ìñigo et al. [16] established 18 WBC/µL and 97 BAC/µL as cut-off points and reported an NPV of 98%. However, others only considered bacteriuria for UTI screening, because the addition of WBC count did not improve diagnostic accuracy [913].
Despite the different cut-off values used in previous studies evaluating automated urine sediment analyzers, the diagnostic accuracy in terms of sensitivity and NPV was always very high (96–99% and 98–99%, respectively), while specificity was low. Specificity was reported to be 59% by Falbo et al. [9] and Martìnez et al. [10], and 75% by Tessari et al. [3] using sediMAX [39] and UriSed (77 Electronika, Busapest, Hungary) [10]. In our study, specificity was 68.4%, which is slightly lower than that obtained by Kim et al. (73.1%) [12], who used the same gold standard. However, the sensitivity of UTI screening tests is far more important than specificity, because all positive samples will be cultured, and false-positive results will not be reported.
Unlike Kim et al. [12], we determined gender-specific cut-off values of BAC and WBC for cobas u 701 and analyzed age distribution. We found better performance in men, where a very high NPV was obtained, and there was a greater reduction in the number of cultures needed and a very low false-negative rate. Overall, the performance of cobas u 701 was very good for subjects ≥20 years. In patients aged 14–19 years, however, diagnostic accuracy tended to be lower, although the small sample size precludes drawing conclusions on the real performance of the analyzer in this age group. Further, our finding that SEC count was a poor predictor of urine culture contamination even if not stratified by sex or age, is in line with other research [1920].
A limitation of our study was that we excluded samples from hospitalized patients, children, and pregnant women because the criteria for culture positivity may be different in these populations, and suboptimal performance has been reported for inpatients [3]. However, the prevalence of positive urine samples and the distribution of causative agents identified in our community setting are in good agreement with the literature [7912]. Therefore, we assume that our samples represent the heterogeneity of urine samples routinely analyzed in clinical microbiology laboratories.
An advantage of automated microscopy analyzer is the substantial reduction in the number of urine samples requiring culture (57% in our study). Previous studies using different systems have reported similar results. For the sediMAX analyzer, a reduction of 54% was reported by Tessari et al. [3], 46.3% by Íñigo et al. [16], and 47% by Falbo et al. [89]. For the iQ200 system, Stürenburg et al. [14] reported a reduction of 35% and figures ranging from 52% to 64.5% have been documented for flow cytometry [713].
In conclusion, cobas u 701 is a useful tool for screening urine samples. The analyzer performed very well in outpatients, except perhaps younger outpatients. After identifying the optimal cut-off points, we could significantly reduce the number of cultures, with a low false-negative rate. Consequently, cobas u 701 can be reliably used to reduce the number of unnecessary urine cultures, at least in outpatients.
Acknowledgments
Roche Diagnostics supplied cobas u 701 and its disposable cuvettes. The sponsor had no role in the study design, data collection and analysis, decision to publish, or writing of manuscript.
Notes
Authors' Disclosures of Potential Conflicts of Interest: The authors declare that they have no conflict of interests.
Author Contributions:
Research conception and design: Ortiz de la Tabla V.
Data acquisition: Gázquez G, Infante A, Martín C, Buñuel F.
Data analysis and interpretation: Ortiz de la Tabla V, Gázquez G, Infante A, Gutiérrez F.
Statistical analysis: Ortiz de la Tabla V, Gutiérrez F.
Drafting of the manuscript: Ortiz de la Tabla V.
Critical revision of the manuscript: Gázquez G, Infante A, Gutiérrez F.
Approval of final manuscript: all authors.
References
1. Stalenhoef JE, van Dissel JT, van Nieuwkoop C. Febrile urinary tract infection in the emergency room. Curr Opin Infect Dis. 2015; 28:106–111. PMID: 25402776.
2. Okada H, Sakai Y, Miyazaki S, Arakawa S, Hamaguchi Y, Kamidono S. Detection of significant bacteriuria by automated urinalysis using flow cytometry. J Clin Microbiol. 2000; 38:2870–2872. PMID: 10921941.
3. Tessari A, Osti N, Scarin M. Screening of presumptive urinary tract infections by the automated urine sediment analyser sediMAX. Clin Chem Lab Med. 2015; 53(S2):s1503–s1508. PMID: 26509783.
4. Brilha S, Proença H, Cristino JM, Hänscheid T. Use of flow cytometry (Sysmex UF-100) to screen for positive urine cultures: in search for the ideal cut-off. Clin Chem Lab Med. 2010; 48:289–292. PMID: 19961394.
5. Evans R, Davidson MM, Sim LR, Hay AJ. Testing by Sysmex UF-100 flow cytometer and with bacterial culture in a diagnostic laboratory: a comparison. J Clin Pathol. 2006; 59:661–662. PMID: 16731608.
6. Zaman Z, Roggeman S, Verhaegen J. Unsatisfactory performance of flow cytometer UF-100 and urine strips in predicting outcome of urine cultures. J Clin Microbiol. 2001; 39:4169–4171. PMID: 11682551.
7. Jolkkonen S, Paattiniemi EL, Kärpänoja P, Sarkkinen H. Screening of urine samples by flow cytometry reduces the need for culture. J Clin Microbiol. 2010; 48:3117–3121. PMID: 20592157.
8. Oyaert M, Delanghe J. Progress in automated urinalysis. Ann Lab Med. 2019; 39:15–22. PMID: 30215225.
9. Falbo R, Sala MR, Signorelli S, Venturi N, Signorini S, Brambilla P. Bacteriuria screening by automated whole-field-image-based microscopy reduces the number of urine cultures. J Clin Microbiol. 2012; 50:1427–1429. PMID: 22238436.
10. Martinez MH, Bottini PV, Levy CE, Garlipp CR. UriSed as a screening tool for presumptive diagnosis of urinary tract infection. Clin Chim Acta. 2013; 425:77–79. PMID: 23906796.
11. Lee W, Ha JS, Ryoo NH. Comparison of the automated cobas u 701 urine microscopy and UF-1000i flow cytometry systems and manual microscopy in the examination of urine sediments. J Clin Lab Anal. 2016; 30:663–671. PMID: 26842372.
12. Kim SH, Song SA, Urm SH, Kook JK, Kim HR, Yong D, et al. Evaluation of the Cobas u 701 microscopy analyser compared with urine culture in screening for urinary tract infection. J Med Microbiol. 2017; 66:1110–1113.
13. Broeren MA, Bahçeci S, Vader HL, Arents NL. Screening for urinary tract infection with the SysmexUF-1000i urine flow cytometer. J Clin Microbiol. 2011; 49:1025–1029. PMID: 21248088.
14. Stürenburg E, Kramer J, Schön G, Cachovan G, Sobottka I. Detection of significant bacteriuria by use of the iQ200 automated urine microscope. J Clin Microbiol. 2014; 52:2855–2860. PMID: 24871218.
15. Kim H, Kim HR, Kim TH, Lee MK. Age-specific cutoffs of the Sysmex UF-1000i automated urine analyzer for rapid screening of urinary tract infections in outpatients. Ann Lab Med. 2019; 39:322–326. PMID: 30623625.
16. Íñigo M, Coello A, Fernández-Rivas G, Carrasco M, Marcó C, Fernández A, et al. Evaluation of the SediMax automated microscopy sediment analyzer and the Sysmex UF-1000i flow cytometer as screening tools to rule out negative urinary tract infections. Clin Chim Acta. 2016; 456:31–35. PMID: 26921459.
17. De Rosa R, Grosso S, Bruschetta G, Avolio M, Stano P, Modolo ML, et al. Evaluation of the Sysmex UF1000i flow cytometer for ruling out bacterial urinary tract infection. Clin Chim Acta. 2010; 411:1137–1142. PMID: 20359474.
18. Manoni F, Tinello A, Fornasiero L, Hoffer P, Temporin V, Valverde S, et al. Urine particle evaluation: a comparison between the UF-1000i and quantitative microscopy. Clin Chem Lab Med. 2010; 48:1107–1111. PMID: 20482296.
19. Mohr NM, Harland KK, Crabb V, Mutnick R, Baumgartner D, Spinosi S, et al. Urinary squamous epithelial cells do not accurately predict urine culture contamination, but may predict urinalysis performance in predicting bacteriuria. Acad Emerg Med. 2016; 23:323–330. PMID: 26782662.
20. Walter FG, Gibly RL, Knopp RK, Roe DJ. Squamous cells as predictors of bacterial contamination in urine samples. Ann Emerg Med. 1998; 31:455–458. PMID: 9546013.
Table 1
Sensitivity, specificity, PPV, and NPV of cobas u 701 for single and combined cut-off points (N=1,360 samples)
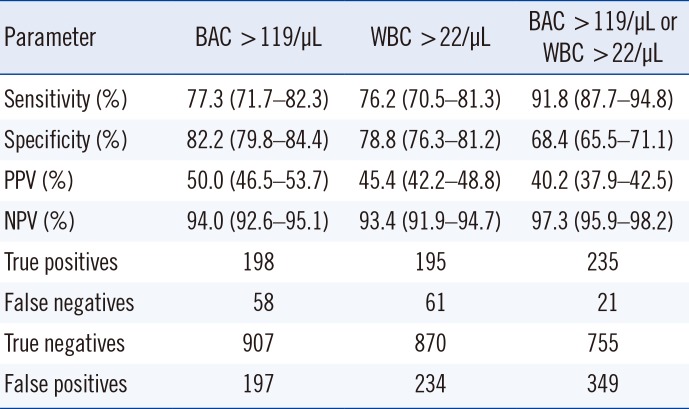
Table 2
Sensitivity, specificity, PPV, and NPV of cobas u 701 according to age categories for combined cut-off points (>119 BAC/µL and >22 WBC/µL; N=1,360 samples)
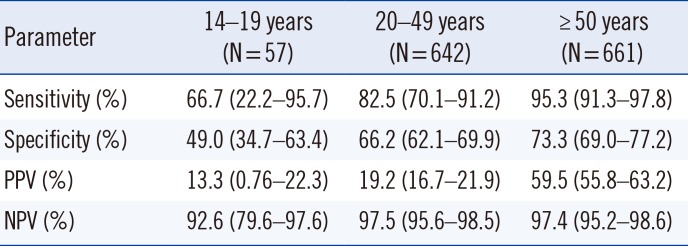
Table 3
Sensitivity, specificity, PPV, and NPV of cobas u 701 in women and men (N=1,360 samples) for single and combined cut-off points




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download