Abstract
Curcumin, an active ingredient of Curcuma longa L., can reduce the concentration of low-density lipoproteins in plasma, in different ways. We had first reported that curcumin exhibits hypocholesterolemic properties by improving the apolipoprotein B (apoB) mRNA editing in primary rat hepatocytes. However, the role of curcumin in the regulation of apoB mRNA editing is not clear. Thus, we investigated the effect of curcumin on the expression of multiple editing components of apoB mRNA cytidine deamination to uridine (C-to-U) editosome. Our results demonstrated that treatment with 50 µM curcumin markedly increased the amount of edited apoB mRNA in primary mouse hepatocytes from 5.13%–8.05% to 27.63%–35.61%, and significantly elevated the levels of the core components apoB editing catalytic polypeptide-1 (APOBEC-1), apobec-1 complementation factor (ACF), and RNA-binding-motif-protein-47 (RBM47), as well as suppressed the level of the inhibitory component glycine-arginine-tyrosine-rich RNA binding protein. Moreover, the increased apoB RNA editing by 50 µM curcumin was significantly reduced by siRNA-mediated APOBEC-1, ACF, and RBM47 knockdown. These findings suggest that curcumin modulates apoB mRNA editing by coordinating the multiple editing components of the editosome in primary hepatocytes. Our data provided evidence for curcumin to be used therapeutically to prevent atherosclerosis.
RNA editing is a unique post-transcriptional modification that makes a mature RNA sequence different from its template DNA sequence by inserting, deleting, or substituting bases [123]. Because RNA editing alters the cellular fate of RNA molecules and their sequence relative to the genome, it plays a critical role in many biological processes [456789]. In mammals, the most common forms of RNA editing are adenosine deamination to inosine and cytidine deamination to uridine (C-to-U). Deficiency in any of the forms of RNA editing can lead to physiological defects or metabolic disorders [456789].
The best characterized C-to-U editing event in mammals occurs in the apolipoprotein B (apoB) mRNA. The C residue at position 6666 of the apoB mRNA is substituted with a U that changes a glutamine codon (CAA) to a termination codon (UAA), generating a truncated protein, apoB-48 [10]. This editing event has important effects on lipoprotein metabolism because apoB plays a central role in the assembly, secretion, and metabolism of low-density lipoproteins (LDLs). ApoB mRNA translates to apoB-100 protein in the absence of C6666RNA editing, and is virtually the only protein component of LDLs that contains the low-density lipoprotein receptor-binding domain; it binds with low-density lipoprotein receptor (LDLR), and mediates the metabolism of LDLs [10]. Due to C6666RNA editing, apoB-48 has only the N-terminal half of apoB-100, and lacks the domain that mediates interaction with the LDLR. Therefore, lipoprotein particles containing apoB-48 are directed to a different metabolic pathway. They possess multiple copies of apolipoprotein E, which assists in the clearance of lipoprotein, through LDLR and LDL receptor-related proteins [1112]. Actually, lipoprotein particles containing apoB-48 clear from plasma more rapidly than those containing apoB-100 (10 min compared to 3 days) [13]. In theory, if it were possible to manipulate or mimic apoB mRNA 6666 C-to-U editing, the secretion of lipoproteins containing apoB-48 could be upregulated, lipoproteins would undergo rapid catabolic clearance, thus reducing the risk of lipid metabolism disorders [14].
Curcumin, an extract from the traditional Chinese medicinal herb Curcuma longa L., possesses numerous important biological activities, including hypocholesterolemic properties [15]. A number of studies have shown that curcumin significantly reduces plasma LDL-cholesterol and improves lipid metabolic disorders [1617]. Liang et al.[18] found that curcumin could significantly reduce the expression of apoB-100 mRNA in live nonalcoholic fatty liver disease mouse, thereby regulating their lipid metabolism. In our previous study, we found that 15 M curcumin raised the level of apoB-48 and reduced the level of apoB-100 by increasing the expression of apoB editing catalytic polypeptide-1 (APOBEC-1) in primary rat hepatocytes [19]. However, apoB mRNA editing is mediated by a coordinated modulation of multiple enzyme components that includes not only APOBEC-1, but also apobec-1 complementation factor (ACF) [20] and RNA-binding-motif-protein-47 (RBM47) [21]. Other auxiliary proteins, including CUG binding protein-2 (CUGBP2), glycine-arginine-tyrosine-rich RNA binding protein (GRY-RBP), heterogenous nuclear ribonucleoprotein (hnRNP)-C1, and apobec-1 binding protein-2 (ABBP2), also participate in this editing event [22]. In this study, we used primary mouse hepatocytes as a model system to investigate the change in APOBEC-1, ACF, RBM47, CUGBP2, GRY-RBP, hnRNP-C1, and ABBP2 expression levels after treatment with curcumin to further clarify the specific functional role of each editing enzyme component in the apoB mRNA editing event.
Institute of Cancer Research (ICR) mice, within 24 h of birth, were purchased from Shanghai Snooker Company (Shanghai, China). Animal experimental procedures were performed in conformity with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication no. 85-23, revised 1996) and were approved by the Ethics Committee of the Animal Laboratory Research Centre of Zhejiang Chinese Medical University (Zhejiang, China; approval no. SYXK [Zhe] 2008-0115). Primary hepatocytes were isolated from ICR mice livers as follows: ICR mice were decapitated and the liver tissues were extracted and placed in cold D-Hanks solution. Liver capsules were peeled off with forceps, and the livers were shredded directly with ophthalmic scissors. Tissues were thoroughly washed and then digested with 0.2% trypsin containing ethylenediaminetetraacetic acid at 37℃ with intermittent shaking. After discarding the supernatant, Dulbecco's modified Eagle's media (DMEM) containing 0.2% type II collagenase was added followed by digestion for 20 min at 37℃. The reaction was terminated by adding DMEM supplemented with 10% fetal bovine serum (FBS); the dispersed cells were filtered through a 200-mesh sieve and centrifuged for 5 min at 800 g. The isolated hepatocytes were washed twice with fresh DMEM and incubated at 37℃ in a humidified atmosphere of 5% CO2. After removing the cultured impurities, the first to third generation cells were used for the subsequent experiments. Cell viability, determined using trypan blue staining, was > 95%.
Primary hepatocytes were seeded in 25 cm2 culture flasks and cultured in DMEM supplemented with 10% FBS. Following a 48 h incubation, the medium was replenished every 24 h, hereafter. Cells (5 × 104 cells/ml) were plated onto glass cover slips in 24-well culture plates and grown for 24 h. They were washed three times with phosphate buffer saline (PBS; pH 7.4), then fixed with fixation solution for 10 min, and permeabilized with 0.1% TritonX-100 for 20 min. Thereafter, the cells were stained using a periodic acid-Schiff kit according to the manufacturer's instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Finally, the stained cells were mounted on glass slides and photographed using a Nikon Eclipse 80i microscope connected to a DS-5M-L1 (Nikon, Melville, NY, USA) camera.
The effect of curcumin on cell viability was determined by MTT assay. Briefly, when the cells reached sub-confluence, they were seeded at a density of 5 × 103 cells/well in 96-well plate and treated with increasing doses of curcumin prepared in DMEM containing 10% FBS. MTT solution (0.5 mg/ml) was added to each well and incubated for 4 h at 37℃. The medium was removed; formazan crystals formed inside the cells were dissolved in 150 µl dimethyl sulfoxide, and then gently shaken for 15 min. The absorbance of MTT-formazan was measured at 570 nm using a SpectraMaxM3 microplate reader (MolecularDevices, Sunnyvale, CA, USA).
Total RNA was isolated from the primary hepatocytes using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Equal quantities (5.0 µg) of total RNA were used for complementary DNA (cDNA) synthesis using the HiFi-MMLV cDNA kits (Beijing Kang Century Biotechnology Co., Ltd., Beijing, China).Ultra SYBR mixture (Beijing Kang Century Biotechnology Co.,Ltd.) was used to perform qRT-PCR. Relative expression of the target gene was normalized against β-actin and analyzed via the 2−ΔΔCt method. All PCR-specific forward and reverse primers were synthesized by GenScript Co., Ltd. (Nanjing, China) (Table 1).
Primary hepatocytes were washed once with PBS and dissolved in Protein Extraction Reagent (Boster Bioengineering, Wuhan, China) containing 1 mM phenylmethanesulfonyl fluoride (PMSF; Roche Molecular Biochemicals, Indianapolis, IN, USA). Total protein was electrophoretically separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transfer to polyvinylidene difluoride (PVDF) membrane. The PVDF membrane was blocked with 1% bovine serum albumin for 2 h at room temperature, and then incubated with the antibodies, separately, overnight at 4℃. The antibody against β-actin was purchased from Hua Bio (Hangzhou HuaAn Biotechnology Co., Ltd., Hangzhou, China), antibodies against APOBEC-1, CUGBP2, and (hnRNP)-C1 were from Santa Cruz (Santa Cruz Biotechnology, Dallas, TX, USA), antibody against ACF (Thermo Fisher Scientific, Waltham, MA, USA), RBM47, and ABBP2 (Proteintech Group, Inc., Chicago, IL, USA), GRY-RBP (Abcam, Cambridge, UK). The membrane was then washed with tris buffered saline-tween solution three times followed by incubation with anti-rabbit immunoglobulin G (IgG) or anti-mouse IgG horseradish peroxidase secondary antibody (Hangzhou HuaAn Biotechnology Co., Ltd.) for 2 h at room temperature. Finally, the protein bands were detected by enhanced chemiluminescence (ECL) detection kit (Amersham Biosciences, Piscataway, NJ, USA). Relative protein expression levels were quantified by using Image J software and normalized to β-actin.
Sequencing of apoBC6666 sites was performed by the direct sequencing method to estimate the editing frequency (Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., Shanghai, China). Briefly, cDNA from three independent biological replicates was PCR amplified using Taq DNA Polymerase (TaKaRa, Otsu, Japan); the primers are listed in Table 1. The PCR product was purified using the AxyPrep PCR cleanup Kit (Axygen Scientific Inc., Union City, CA, USA) and sequenced using the forward primer. In parallel, we also extracted the hepatocyte genome and PCR amplified apoB gene fragments as a control. The ratio between the C and T peak height in individual chromatograms was measured using FinchTV (https://digitalworldbiology.com/FinchTV). Percent editing was calculated as the T peak height relative to the total peak height [T / (C + T) × 100].
Lipofectamine 2000 (Invitrogen, Life Technologies, Carlsbad, CA, USA) was used to introduce APOBEC-1 siRNA (MSS202127), ACF siRNA (MSS245486), and RBM47 siRNA (MSS205172) oligonucleotides (Invitrogen, Life Technologies) into primary hepatocytes according to the manufacturer's instructions. A scrambled siRNA was used as the negative control. After 48 h incubation, total RNA was isolated from the transfected primary hepatocytes and qRT-PCR was performed using specific primers (Table 1) to evaluate the effect of siRNA-mediated gene knockdown.
Data are expressed as mean ± standard deviation. Statistical analysis was performed using one-way analysis of variance for multiple comparisons and t-tests among groups. The data were analyzed using IBM SPSS Statistics 21.0 (IBM, Armonk, NY, USA). Statistical significance was considered if p < 0.05.
Hepatocytes, isolated immediately from newborn mouse liver had a rounded cell body with a translucent cytoplasm. After overnight incubation, the cells appeared more polygonal in shape (Fig. 1A). Healthy hepatocytes were devoid of granulations or blebs. To identify the hepatocytes, we performed periodic acid-Schiff staining. As shown in Fig. 1B, dense pink glycogen granules were observed in the cytoplasm of the cells, around the nuclei.
The MTT assay was used to determine the dose range of curcumin that does not affect the proliferation of primary mouse hepatocytes. As shown in Fig. 2A, the cell viability was not significantly repressed by treatment with < 50 µM dose of curcumin for 24 h, but treatment with 60 µM curcumin resulted in an approximate 40% decrease in cell survival. Therefore, three concentrations (5, 25, and 50 µM) were selected to further assess the cytotoxicity of curcumin after 36 h and 48 h of treatment. Results showed no decrease in viability between the control (0 µM) and curcumin-treated (5, 25, and 50 µM) cells after 36 h and 48 h (Fig. 2B). Therefore, 5, 25, and 50 µM curcumin were considered the optimum concentration for subsequent experiments.
To determine how curcumin affects the ability of primary mouse hepatocytes to edit apoB mRNA, total cellular RNA was isolated from primary hepatocytes after treatment with 5, 25, and 50 µM curcumin for 48 h and then assayed for apoB mRNA editing efficiency as described in materials and methods. As shown in Fig. 3, the C-to-U editing signal was detected in all the RT-PCR sequencing groups, but not in the genome which was isolated from 50 µM curcumin treated hepatocytes. The editing efficiency in isolated hepatocytes varied with each individual mouse and ranged from 5.13%–8.05% (n = 7); after treatment with 5 and 25 µM curcumin, the editing efficiency demonstrated no significant changes (4.48%–8.62%, 5.06%–8.41%, respectively). However, increasing the concentration of curcumin to 50 µM, enhanced the editing efficiency of apoB mRNA C6666 site (27.63%–35.61%), indicating that treatment with an intermediate concentration of curcumin increased the amount of apoB-48 and reduced the amount of apoB-100 in cultured primary mouse hepatocytes.
To further explore the mechanism of curcumin in upregulating apoB mRNA editing, we evaluated the change in the expression of the multiple enzyme components following curcumin treatment, by qRT-PCR and western blotting. As shown in Fig. 4A, the mRNA level of APOBEC-1 was approximately four-fold higher in cells treated with 50 µM curcumin than in untreated, 5, and 25 µM curcumin-treated cells. The mRNA levels of RBM47 and ACF were also upregulated by 50 µM curcumin treatment. Interestingly, 25 µM curcumin treatment increased the mRNA level of ACF as well. Furthermore, we found that treatment with 50 µM curcumin significantly reduced the mRNA level of GRY-RBP, an inhibitory component of C-to-U RNA editing enzyme complex, while curcumin treatment had no effect on other auxiliary proteins CUGBP2 and hnRNP-C1. In addition, protein expression was consistent with the mRNA status (Fig. 4B). Next, APOBEC-1, ACF, and RBM47 siRNA oligonucleotides were transfected into primary hepatocytes to evaluate whether the increased apoB RNA editing by 50 µM curcumin disappeared when the expression of the editosome components was knockdowned. As shown in Fig. 5A, both in untreated and 50 µM curcumin treated group, the expression of APOBEC-1, ACF, and RBM47 was decreased in siRNA-mediated gene knockdown cells compared with siRNA NC transfected and untransfected cells. As anticipated, the editing efficiency of apoB mRNA C6666 site in APOBEC-1, ACF, and RBM47 knockdown hepatocytes was significantly decreased, only had 1.47%–2.62% (Fig. 5B). In 50 µM curcumin treated group, the editing efficiency in APOBEC-1, ACF, and RBM47 knockdown hepatocytes was also reduced (8.43%–16.27%) compared with siRNA NC transfected and untransfected hepatocytes (Fig. 5B). Collectively, these results suggested that curcumin affected apoB mRNA editing efficiency by regulating the expression of the components of the editing enzyme complex.
ApoB mRNA editing is an exquisitely precise nuclear process that alters a single nucleotide in a molecule containing over 14,000 bases. This editing event requires the assembly of a multiprotein editosome. Among them, APOBEC-1, which has cytidine deaminase activity, is the catalytic component of the editing enzyme complex [23]. ACF (recently renamed A1CF) is an RNA-binding protein that serves as the RNA recognition component of the editing enzyme [2425], while RBM47 is also involved in the editing events via interaction with APOBEC-1 and ACF [21]. Besides the afore mentioned core components, the editosome also includes other auxiliary inhibitory proteins such as CUGBP2, GRY-RBP, hnRNP-C1, and ABBP2 [22262728].
Studies have indicated that apoB mRNA editing is tissue-specific [293031] and modulated by several interventions (e.g. insulin, ethanol), which could regulate the expression of APOBEC-1 or the auxiliary proteins [323334353637]. Because there are differences in the apoB100- and apoB48-containing nascent particles in their dependence on a second step of lipidation [38], and the rate of clearance of their respective remnants from the circulation, the degree of apoB mRNA editing has important physiological implications.
Curcumin has been shown to regulate the lipid mechanism in hepatocytes [3940] as well as the lipoprotein concentrations in serum [4142]. Our previous study showed that intermediate concentration of curcumin increased the degree of apoB mRNA editing in primary murine hepatocytes by up-regulating the expression of APOBEC-1. The current study aimed to clarify the effect of curcumin on other components of editosome. As shown in Fig. 4, in primary mouse hepatocytes, 50 µM curcumin significantly increased the mRNA and protein levels of APOBEC-1, ACF, and RBM47, but decreased the levels of GRY-RBP. No effect of curcumin was observed on inhibitory proteins CUGBP2, hnRNP-C1, and ABBP2. In addition, the increased apoB RNA editing by 50 µM curcumin was significantly reduced by siRNA-mediated knockdown of APOBEC-1, ACF, and RBM47. These results suggested that the intermediate concentration of curcumin enhances the apoB mRNA editing efficiency by regulating the expression of all the three core components, but not the auxiliary inhibitory members in editing enzyme complex (Fig. 6); there is a coordinated modulation among these four components in vivo. The increased, and similar expression of ACF at 25 µM and 50 µM curcumin, suggests that curcumin does not affect apoB mRNA editing in a dose-dependent manner. Researches have shown that curcumin increased the expression of CUGBP2 in pancreatic cancer cells and inhibited pancreatic tumor growth via the CUGBP2-mediated post-transcriptional regulation pathway [4344]. However, we didn't detect the influence of 5, 25, and 50 µM curcumin on the mRNA level of CUGBP2 in primary mouse hepatocytes. Curcumin has been reported to possess antitumor properties but does not cause any damage to the normal cells, and downregulated the expression of bcl-2 in two human breast adenocarcinoma cell lines but had no significant effect on bcl-2 expression in normal rat hepatocytes [45]. So we believed that the effect of curcumin on gene expression in normal cells was different with that in malignant cells.
Van Mater et al. [46] reported that apoB mRNA editing efficiency in isolated, primary rat hepatocytes ranged from 38% to 58% and varied with each individual rat, but the proportion of edited apoB mRNA in our primary mouse hepatocytes was much lower, ranging from 5.13% to 8.05%. It is worth noting that Van Mater et al. [46] used hepatocytes at the time of their preparation, before they were plated. In our study, primary hepatocytes were treated with curcumin for 48 h, and when they were used to assay for apoB mRNA editing efficiency, it was more than 96 h from the time of their isolation. In addition, Van Mater et al. [46] isolated hepatocytes from rat, and we isolated hepatocytes from mouse. Accordingly, we considered that the differences in the data may be due to tissue-specific expressions and nutritional perturbations.
Unlike the dramatic change in apoB mRNA editing efficiency that is apparent with adenoviral delivery of exogenous APOBEC-1 [47], curcumin-induced changes in the apoB mRNA editing efficiency were comparatively small, because curcumininduced editing events involved the coordinated expression of multiple editing components in vivo. In conclusion, our findings throw a new light on the hypolipidemic effect of curcumin that elevates the level of apoB-48 and reduces the level of apoB-100 by coordinating the expression of some components of the editosome; curcumin may be used therapeutically to reduce levels of atherogenic lipoproteins and prevent atherosclerosis.
ACKNOWLEDGEMENTS
This study was supported in part by research fund for National Natural Science Foundation of China (81602624), Research Fund Project of Zhejiang Chinese Medical University (2015ZZ14).
Notes
References
1. Covello PS, Gray MW. On the evolution of RNA editing. Trends Genet. 1993; 9:265–268. PMID: 8379005.

2. Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006; 7:919–931. PMID: 17139332.

3. Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010; 26:221–230. PMID: 20395010.

4. Qin YR, Qiao JJ, Chan TH, Zhu YH, Li FF, Liu H, Fei J, Li Y, Guan XY, Chen L. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014; 74:840–851. PMID: 24302582.

5. Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, Liu M, Yuan YF, Fu L, Kong KL, Qi L, Li Y, Zhang N, Tong AH, Kwong DL, Man K, Lo CM, Lok S, Tenen DG, Guan XY. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013; 19:209–216. PMID: 23291631.

6. Shtrichman R, Germanguz I, Mandel R, Ziskind A, Nahor I, Safran M, Osenberg S, Sherf O, Rechavi G, Itskovitz-Eldor J. Altered A-to-I RNA editing in human embryogenesis. PLoS One. 2012; 7:e41576. PMID: 22859999.

7. Hartner JC, Schmittwolf C, Kispert A, Müller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004; 279:4894–4902. PMID: 14615479.

8. Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015; 349:1115–1120. PMID: 26275108.

9. Anant S, Murmu N, Houchen CW, Mukhopadhyay D, Riehl TE, Young SG, Morrison AR, Stenson WF, Davidson NO. Apobec-1 protects intestine from radiation injury through posttranscriptional regulation of cyclooxygenase-2 expression. Gastroenterology. 2004; 127:1139–1149. PMID: 15480992.

10. Anant S, Blanc V, Davidson NO. Molecular regulation, evolutionary, and functional adaptations associated with C to U editing of mammalian apolipoproteinB mRNA. Prog Nucleic Acid Res Mol Biol. 2003; 75:1–41. PMID: 14604008.

11. Anant S, Davidson NO. Molecular mechanisms of apolipoprotein B mRNA editing. Curr Opin Lipidol. 2001; 12:159–165. PMID: 11264987.

12. Pitas RE, Innerarity TL, Mahley RW. Cell surface receptor binding of phospholipid XMLLink_XYZ protein complexes containing different ratios of receptor-active and -inactive E apoprotein. J Biol Chem. 1980; 255:5454–5460. PMID: 7372644.
13. Innerarity TL, Mahley RW. Enhanced binding by cultured human fibroblasts of apo-E-containing lipoproteins as compared with low density lipoproteins. Biochemistry. 1978; 17:1440–1447. PMID: 206278.

14. Castelli WP, Anderson K, Wilson PW, Levy D. Lipids and risk of coronaryheart disease. The Framingham Study. Ann Epidemiol. 1992; 2:23–28. PMID: 1342260.
15. Sandur SK, Ichikawa H, Pandey MK, Kunnumakkara AB, Sung B, SethiG , Aggarwal BB. Role of pro-oxidants and antioxidants in the anti-inflammatory and apoptotic effects of curcumin (diferuloylmethane). Free Radic Biol Med. 2007; 43:568–580. PMID: 17640567.

16. Dou X, Fan C, Wo L, Yan J, Qian Y, Wo X. Curcumin up-regulates LDL receptor expression via the sterol regulatory element pathway in HepG2 cells. Planta Med. 2008; 74:1374–1379. PMID: 18704882.

17. Peschel D, Koerting R, Nass N. Curcumin induces changes in expression of genes involved in cholesterol homeostasis. J Nutr Biochem. 2007; 18:113–119. PMID: 16713233.

18. Liang YF, Zhang DD, Sun LL, Ni L, Liu JX, Wang FF, Fang YC, Wang SQ, Ma XR, Liu L, Wu JM. Research on the effect of Curcumin and Ganoderma lucidum on the expression of ApoB100 mRNA in NAFLD. Heilongjiang Med Pharm. 2013; 36:39–40.
19. Tian N, Li X, Luo Y, Han Z, Li Z, Fan C. Curcumin regulates the metabolism of low density lipoproteins by improving the C-to-U RNA editing efficiency of apolipoprotein B in primary rat hepatocytes. Mol Med Rep. 2014; 9:132–136. PMID: 24173373.

20. Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, Davidson NO. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J Biol Chem. 2001; 276:47338–47351. PMID: 11577082.
21. Fossat N, Tourle K, Radziewic T, Barratt K, Leibhold D, Studdert JB, Power M, Jones V, Loebel DA, Tam PP. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014; 15:903–910. PMID: 24916387.
22. Chen Z, Eggerman TL, Patterson AP. ApoB mRNA editing is mediated by a coordinated modulation of multiple apoB mRNA editing enzyme components. Am J Physiol Gastrointest Liver Physiol. 2007; 292:G53–G65. PMID: 16920700.

23. Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993; 260:1816–1819. PMID: 8511591.

24. Lellek H, Kirsten R, Diehl I, Apostel F, Buck F, Greeve J. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J Biol Chem. 2000; 275:19848–19856. PMID: 10781591.

25. Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol. 2000; 20:1846–1854. PMID: 10669759.

26. Fossat N, Tam PP. Re-editing the paradigm of Cytidine (C) to Uridine (U) RNA editing. RNA Biol. 2014; 11:1233–1237. PMID: 25585043.

27. Chen Z, Eggerman TL, Bocharov AV, Baranova IN, Vishnyakova TG, Kurlander RJ, Csako G, Patterson AP. Hypermutation of ApoB mRNA by rat APOBEC-1 overexpression mimics APOBEC-3 hypermutation. J Mol Biol. 2012; 418:65–81. PMID: 22326345.

28. Chen Z, Eggerman TL, Bocharov AV, Baranova IN, Vishnyakova TG, Csako G, Patterson AP. Hypermutation induced by APOBEC-1 overexpression can be eliminated. RNA. 2010; 16:1040–1052. PMID: 20348446.

29. Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, Silberman SR, Cai SJ, Deslypere JP, Rosseneu M, Gotto M, Li WH, Chan L. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987; 238:363–366. PMID: 3659919.

30. Greeve J, Altkemper I, Dieterich JH, Greten H, Windler E. Apolipoprotein B mRNA editing in 12 different mammalian species: hepatic expression is reflected in low concentrations of apoB-containing plasma lipoproteins. J Lipid Res. 1993; 34:1367–1383. PMID: 8409768.

31. Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987; 50:831–840. PMID: 3621347.

32. Chen Z, Eggerman TL, Patterson AP. Phosphorylation is a regulatory mechanism in apolipoprotein B mRNA editing. Biochem J. 2001; 357:661–672. PMID: 11463337.

33. Chen Z, Eggerman TL, Potosky D, Arborati M, Patterson AP. Calcium increases apolipoprotein B mRNA editing. Biochem Biophys Res Commun. 2000; 277:221–227. PMID: 11027667.

34. Funahashi T, Giannoni F, DePaoli AM, Skarosi SF, Davidson NO. Tissue-specific, developmental and nutritional regulation of the gene encoding the catalytic subunit of the rat apolipoprotein B mRNA editing enzyme: functional role in the modulation of apoB mRNA editing. J Lipid Res. 1995; 36:414–428. PMID: 7775854.

35. Giangreco A, Sowden MP, Mikityansky I, Smith HC. Ethanol stimulates apolipoprotein B mRNA editing in the absence of de novo RNA or protein synthesis. Biochem Biophys Res Commun. 2001; 289:1162–1167. PMID: 11741314.

36. Lindén D, Sjöberg A, Asp L, Carlsson L, Oscarsson J. Direct effects of growth hormone on production and secretion of apolipoprotein B from rat hepatocytes. Am J Physiol Endocrinol Metab. 2000; 279:E1335–E1346. PMID: 11093922.

37. Patterson AP, Chen Z, Rubin DC, Moucadel V, Iovanna JL, Brewer HB Jr, Eggerman TL. Developmental regulation of apolipoprotein B mRNA editing is an autonomous function of small intestine involving homeobox gene Cdx1. J Biol Chem. 2003; 278:7600–7606. PMID: 12493769.

38. Wang Y, McLeod RS, Yao Z. Normal activity of microsomal triglyceride transfer protein is required for the oleate-induced secretion of very low density lipoproteins containing apolipoprotein B from McA-RH7777 cells. J Biol Chem. 1997; 272:12272–12278. PMID: 9139669.

39. Tai MH, Chen PK, Chen PY, Wu MJ, Ho CT, Yen JH. Curcumin enhances cell-surface LDLR level and promotes LDL uptake through downregulation of PCSK9 gene expression in HepG2 cells. Mol Nutr Food Res. 2014; 58:2133–2145. PMID: 25164566.

40. Lu C, Zhang F, Xu W, Wu X, Lian N, Jin H, Chen Q, Chen L, Shao J, Wu L, Lu Y, Zheng S. Curcumin attenuates ethanol-induced hepatic steatosis through modulating Nrf2/FXR signaling in hepatocytes. IUBMB Life. 2015; 67:645–658. PMID: 26305715.

41. Panahi Y, Kianpour P, Mohtashami R, Jafari R, Simental-Mendía LE, Sahebkar A. Curcumin lowers serum lipids and uric acid in subjects with nonalcoholic fatty liver disease: a randomized controlled trial. J Cardiovasc Pharmacol. 2016; 68:223–229. PMID: 27124606.

42. Neerati P, Devde R, Gangi AK. Evaluation of the effect of curcumin capsules on glyburide therapy in patients with type-2 diabetes mellitus. Phytother Res. 2014; 28:1796–1800. PMID: 25044423.

43. Jakstaite A, Maziukiene A, Silkuniene G, Kmieliute K, Dauksa A, Paskauskas S, Gulbinas A, Dambrauskas Z. Upregulation of cugbp2 increases response of pancreatic cancer cells to chemotherapy. Langenbecks Arch Surg. 2016; 401:99–111. PMID: 26691217.

44. Subramaniam D, Ramalingam S, Linehan DC, Dieckgraefe BK, Postier RG, Houchen CW, Jensen RA, Anant S. RNA binding protein CUGBP2/CELF2 mediates curcumin-induced mitotic catastrophe of pancreatic cancer cells. PLoS One. 2011; 6:e16958. PMID: 21347286.

45. Syng-Ai C, Kumari AL, Khar A. Effect of curcumin on normal and tumor cells: role of glutathione and bcl-2. Mol Cancer Ther. 2004; 3:1101–1108. PMID: 15367704.
46. Van Mater D, Sowden MP, Cianci J, Sparks JD, Sparks CE, Ballatori N, Smith HC. Ethanol increases apolipoprotein B mRNA editing in rat primary hepatocytes and McArdle cells. Biochem Biophys Res Commun. 1998; 252:334–339. PMID: 9826530.

47. Teng B, Blumenthal S, Forte T, Navaratnam N, Scott J, Gotto AM Jr, Chan L. Adenovirus-mediated gene transfer of rat apolipoprotein B mRNA-editing protein in mice virtually eliminates apolipoprotein B-100 and normal low density lipoprotein production. J Biol Chem. 1994; 269:29395–29404. PMID: 7961918.

Fig. 1
The morphology of primary mouse hepatocytes and their identification.
(A) The morphology of primary mouse hepatocytes. (B) PAS staining of primary mouse hepatocytes. Magnifications: (A) bar = 20 µm; (B) bar = 10 µm. PAS, periodic acid-Schiff stain.
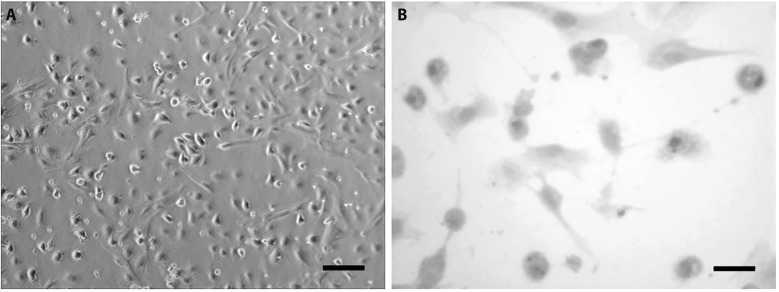
Fig. 2
The viability of primary mouse hepatocytes was determined by the MTT assay.
(A) Cells were exposed to curcumin at different concentrations for 24 h. (B) Cells were exposed to 5, 25, and 50 µM curcumin for different time periods. Each point and vertical bar presents the mean ± standard deviation of three independent experiments. **p < 0.01.

Fig. 3
Comparison of editing efficiency between curcumin treatment groups and control group at the apolipoprotein BC (apoBC)6666 editing site.
Sequencing chromatograms around the editing sites are generated by the direct sequencing method using real-time polymerase chain reaction (RT-PCR) (or PCR) product from the RNA (or genome) was subjected to editing reactions. Various colors correspond to the different nucleotides in the chromatograms: red (thymine), green (adenosine), and blue (cytosine). A black arrowhead indicates the editing sites. The C and T peaks correspond to unedited and edited signals, respectively. **p < 0.01.

Fig. 4
Effect of curcumin on the (A) mRNA and (B) protein level of apolipoprotein B (apoB) mRNA editing enzyme components in primary mouse hepatocytes.
The cells were treated with 5, 25, and 50 µM curcumin. After 48 h treatment, RNA was isolated from cells, real-time quantitative polymerase chain reaction and Western blotting was carried out to evaluate the expression levels of the RNA editing enzymes. Results were repeated twice and are displayed as mean ± standard deviation. APOBEC-1, apoB editing catalytic polypeptide-1; RBM47, RNA-binding-motif-protein-47; ACF, apobec-1 complementation factor; GRY-RBP, glycine-arginine-tyrosine-rich RNA binding protein; hnRNP, heterogenous nuclear ribonucleoprotein; CUGBP2, CUG binding protein-2; ABBP2, apobec-1 binding protein-2. *p < 0.05.
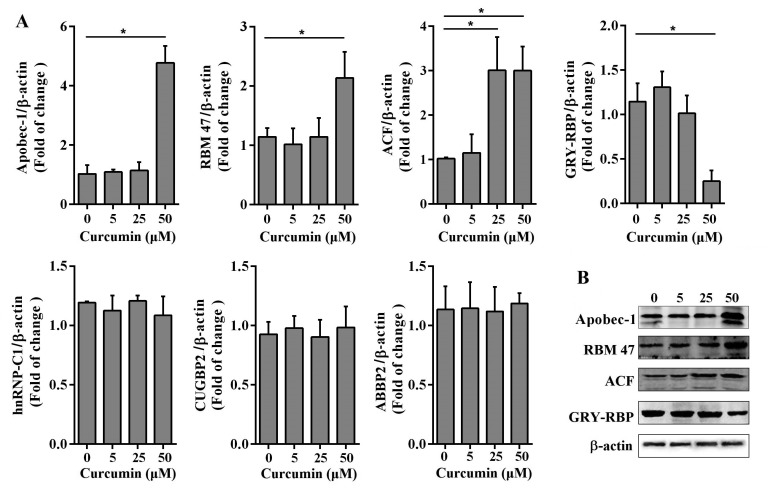
Fig. 5
Effect of siRNA-mediated gene knockdown on apolipoprotein B (apoB) mRNA editing in primary hepatocytes.
(A) siRNA-mediated gene knockdown reduced the expression of APOBEC-1, ACF, and RBM47 in primary hepatocytes. (B) siRNA-mediated gene knockdown reduced the editing efficiency of apoB mRNA C6666 site. Cells were transfected with APOBEC-1, ACF, and RBM47 siRNA oligonucleotides, then untreated or treated with 50 µM curcumin for 48 h. sicon: siRNA negative control. APOBEC-1, apoB editing catalytic polypeptide-1; RBM47, RNA-binding-motif-protein-47; ACF, apobec-1 complementation factor. *p < 0.05, **p < 0.01.
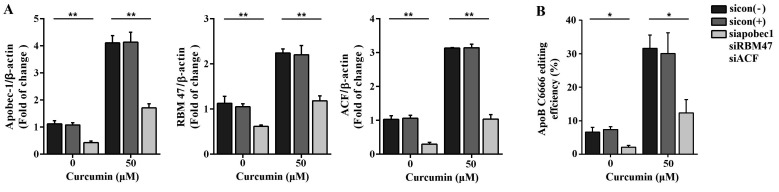
Fig. 6
Curcumin modulates the apolipoprotein B (apoB) mRNA editing by coordinating the expression of multiple editing components of cytidine deamination to uridine (C-to-U) editosome.
Curcumin increased the expression of the three core components APOBEC-1, RBM47, and ACF, but reduced the expression of only one auxiliary inhibitory protein GRY-RBP. APOBEC-1, apoB editing catalytic polypeptide-1; RBM47, RNA-binding-motif-protein-47; ACF, apobec-1 complementation factor; GRY-RBP, glycine-arginine-tyrosine-rich RNA binding protein; hnRNP, heterogenous nuclear ribonucleoprotein; CUGBP2, CUG binding protein-2; ABBP2, apobec-1 binding protein-2; LDLR-BD, low-density lipoprotein receptor-binding domain.
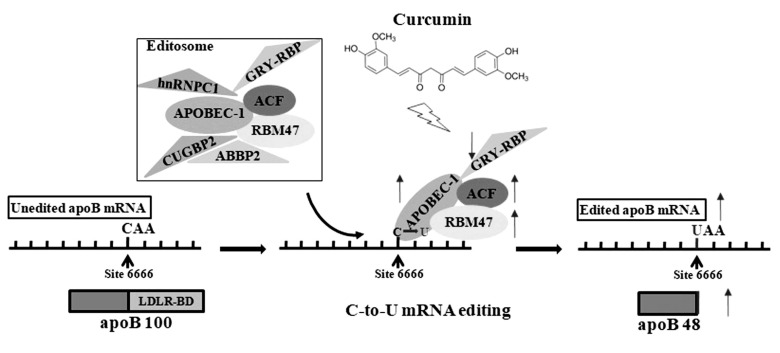
Table 1
Primers used for polymerase chain reaction amplification of mRNA
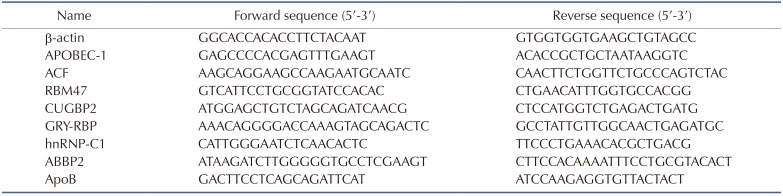
APOBEC-1, apoB editing catalytic polypeptide-1; ACF, apobec-1 complementation factor; RBM47, RNA-binding-motif-protein-47; CUGBP2, CUG binding protein-2; GRY-RBP, glycine-arginine-tyrosine-rich RNA binding protein; hnRNP, heterogenous nuclear ribonucleoprotein; ABBP2, apobec-1 binding protein-2; ApoB, apolipoprotein B.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download