Abstract
Despite increased evidence of bio-activity following far-infrared (FIR) radiation, susceptibility of cell signaling to FIR radiation-induced homeostasis is poorly understood. To observe the effects of FIR radiation, FIR-radiated materials-coated fabric was put on experimental rats or applied to L6 cells, and microarray analysis, quantitative real-time polymerase chain reaction, and wound healing assays were performed. Microarray analysis revealed that messenger RNA expressions of rat muscle were stimulated by FIR radiation in a dose-dependent manner in amount of 10% and 30% materials-coated. In 30% group, 1,473 differentially expressed genes were identified (fold change [FC] > 1.5), and 218 genes were significantly regulated (FC > 1.5 and p < 0.05). Microarray analysis showed that extracellular matrix (ECM)-receptor interaction, focal adhesion, and cell migration-related pathways were significantly stimulated in rat muscle. ECM and platelet-derived growth factor (PDGF)-mediated cell migration-related genes were increased. And, results showed that the relative gene expression of actin beta was increased. FIR radiation also stimulated actin subunit and actin-related genes. We observed that wound healing was certainly promoted by FIR radiation over 48 h in L6 cells. Therefore, we suggest that FIR radiation can penetrate the body and stimulate PDGF-mediated cell migration through ECM-integrin signaling in rats.
Electromagnetic radiation can be divided into different parts based on wavelength. Infrared region lies between microwave and visible radiation, and may be further divided into near-, middle-, and far-infrared (FIR) rays. FIR region refers to radiation wavelengths between 5.6 and 1,000 µm that is associated with health benefits [1].
Similar wavelengths of FIR radiation and vibration frequency characteristics of human body allow FIR heat to penetrate 2.5-cm deep under the skin, to the muscles, blood vessels, lymphatic glands, and nerves [2]. Even low-intensity light significantly influences wound healing [3]. The function of infrared sauna and infrared radiator is based on the vibrational effect of FIR radiation [45]. Vibration frequency required to penetrate the human body wavelengths between 6 and 20 µm [2]. Recent studies have shown that FIR affects, skin microcirculation [678], blood flow recovery [91011], reactive oxygen species reduction [4], endothelial nitric oxide synthase up-regulation [12], neuromuscular recovery [13], and wound healing [10]. FIR radiation has also been shown to cause stimulation of the glutathione-dependent system, including induction of messenger RNA (mRNA) expressions [14]. This proves that mRNA expression is stimulated by FIR radiation alone. A recent study has shown that mid-infrared radiation plays a beneficial role in cancer therapy [15]. Despite increased evidences of bio-activity following FIR radiation, mechanism of cell signaling to FIR radiation-induced health is poorly understood. It would be interesting and important to understand how FIR radiation modulates cell survival pathway and promotes wound healing pathway as infrared therapies increasing, including IR radiators and FIR saunas.
Integrins are trans-membrane receptors that stimulate attachment with components of the extracellular matrix (ECM) during cell migration, and activate the signaling of cell migration pathway molecules to mediate direct interaction with both the cytoskeleton and ECM [16171819]. Platelet-derived growth factor (PDGF) is an important mitotic factor of fibroblasts and one of the several growth factors that regulate cell growth, cell migration, proliferation, blood vessel formation, differentiation, and transformation via ECM-PDGF signaling [122021222324252627]. PDGF induces ERK phosphorylation and promotes cell migration that is specifically PDGF receptor-dependent [2829]. Also, fibronectin has been found to increase with PDGF receptor protein [3031]. Upon integrin-mediated cellular binding to fibronectin via src homology 2 (SH2)-domain-mediated association of the growth factor receptor-bound protein 2 (GRB2) adaptor protein, then GRB2 SH2-domain bind directly to focal adhesion kinase autophosphorylation stimulates alpha-actinin binding [32]. Actin and actin-binding proteins are essential for maintaining cell shape, motility, division, adhesion, and morphogenesis during intracellular protein trafficking in cytoskeleton [33]. Integrin and actin are important physical linkers or scaffolds for cell migration that is regulated by cell retraction and adhesion [3435]. As the neuronal cells move, myosin contributes to control cellular shape and motility, and alpha-actinin regulates the length and density of dendritic spine [33]. Cell migration contributes to tissue repair and regeneration. FIR radiation from a ceramic FIR emitter was reported to promote burn-wound healing via migration and proliferation of keratinocytes [1636].
In this study, exposure to the FIR was provided using FIR radiated fabric, called bio-active materials-coated fabric (BMCF), according to Lee et al. [37]. We examined the effects of FIR on rat skeletal muscle tissue and rat skeletal muscle myocyte (L6). We aimed to suggest a mechanism of action for the health benefits of FIR rays from the FIR-radiated materials-coated fabric.
The fabric samples, provided by Ventex Inc. (Seoul, Korea), contained 30 kinds of minerals, such as SiO2, Mg, Al2O3, Na, Ca, and Fe2O3 that were blended with acrylic resin at a desired ratio and coated on one side of the fabric. Germacolor Inc. (Mespelbrunn, Germany) and Ventex collaborated to develop this fabric, and the clothing made therefrom has a layer that radiates bioactive energy. The bio-active materials radiate unique energy, generated according to the molecular structure and vibration of each element. The clothing made from BMCF was put on rats. The emission rate between 5 and 20 µm was verified by the Korea Far Infrared Association (Seoul, Korea) to be 89.5%. The emission energy was 3.45 × 102 W/m2 × µm, at 37℃ (Issue No. KFI-789). Any kind of unusual skin reaction was not detected upon safety evaluation of BMCF (data not shown).
Sprague-Dawley rats (12-week-old, male), weighing 350 ± 15 g, were obtained from Koatech Inc. (Pyeongtaek, Korea). The animals were allowed to acclimatize for a week before the experiments, in a standard, controlled-environment housing with temperature of 22℃ ± 3℃, humidity of 50% ± 10%, 12 h light-dark cycle, and ventilation 10 to 15 times/h with wind velocity of 10 to 20 cm/sec. The animals were fed filtered tap water and purified pellet diet. The protocols used conformed to the Guide for the Care and Use of Laboratory Animals, National Institutes of Health, and were approved by the Ethics Committee for Laboratory Animals of Chung-Ang University (approval number: 14-0023). The animals were divided into a control group and two experimental groups (three animals in each). Cloth made from conventional fabric was used for the control group while 10% and 30% BMCF were used for each experimental group, respectively.
The Affymetrix Whole-transcript Expression array was performed according to the manufacturer's protocol (GeneChip Whole Transcript PLUS reagent kit; Affymetrix, Santa Clara, CA, USA) in rat sub-trapezial muscle. The sense complementary DNA (cDNA) was then fragmented and biotin-labeled with terminal deoxynucleotidyl transferase using the GeneChip WT Terminal labeling kit (Affymetrix). Approximately 5.5 µg of labeled DNA target was hybridized to the Affymetrix GeneChip Rat 2.0 ST Array (Affymetrix) at 45℃ for 16 h. Hybridized arrays were washed and stained on a GeneChip Fluidics Station 450 and scanned on a GCS3000 Scanner (Affymetrix). Signal values were computed using the Affymetrix® GeneChip™ Command Console software (Affymetrix) [38]. The pathway analysis of differentially expressed genes (DEGs) was based on Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene ontology (GO) databases.
The DEGs of rat sub-trapezial muscle which fold change (FC) > 1.5 and p < 0.05 were analyzed by Search Tool for the Retrieval of Interacting Genes (STRING; www.string-db.org) ver. 10.5 to identify protein, predicted by gene expression, interactions and identified herb genes. Twenty two DEGs of 30% group were represented in the protein-protein interaction network analysis [39].
In order to observe the effects of FIR radiation-induced mRNA expression of cell migration-related proteins in rat sub-trapezial muscle, mRNA expression of PDGF-mediated cell migration-related genes was verified via LightCycler 2.0 (Roche, Basel, Switzerland). Integrin alpha 5 (Itga5), Itga11, matrix metallopeptidase 2 (Mmp2), actin beta (Actb), extracellular matrix protein 1 (Ecm1), Ecm2, PDGF receptor-like (Pdgfrl), platelet-derived growth factor D (Pdgfd), PDGF receptor alpha (Pdgfra), ras homolog family member B (RhoB) and RhoJ expression were measured by Fast Start DNA Master SYBR Green I kit (Roche). All primers used in qRT-PCR analysis were described in Table 1. Gapdh was used as an endogenous control. Each reaction mixture contained 0.8 µl MgCl2 (final concentration 3 mM), 0.2 µl forward primer (final concentration 0.2 µM), 0.2 µl reverse primer (final concentration 0.2 µM), 1 µl LightCycler DNA Master SYBR Green I 10 X, 1 µl cDNA template, and volume made up to 10 µl with distilled water.
L6 cells were cultured in general medium (10% fetal bovine serum [FBS], 1% penicillin/streptomycin in Dulbecco's modified Eagle medium [DMEM]) for 24 h. After incubation, 70 µl of cell suspension at a density of 8.4 × 105 cells per 1 ml was added to Culture-Insert 2 Well (Culture-Inserts 2 Well; Ibidi, Madison, WI, USA). For FIR radiation conditions, 0% and 10% materials-coated fabric were attached inside of the culture dish lid with a distance of 1 cm from the cells. Upon incubation at 37℃ and 5% CO2 for 24 h, the Culture-Insert 2 Well that creates cell-free gap of approximately 500 µm was removed and washed with PBS to remove cell debris and non-attached cells in starvation medium (1% FBS, 1% penicillin/streptomycin in DMEM) was applied for 48 h. Images were taken at 0, 24, and 48 h with a DCM-800 digital camera (ScopeTek, Hangzhou, China) and under a IX-50 microscope (Olympus, Osaka, Japan).
Raw data were extracted automatically in Affymetrix data extraction protocol using the software provided by Affymetrix GeneChip® Command Console®. After importing CEL files, the data were summarized and normalized with robust multi-average (RMA) method, implemented by Affymetrix® Expression Console™ Software. We exported the result with gene-level RMA analysis and performed DEG analysis. Statistical significance of the expression data was determined using local-pooled-error test and FC, in which the null hypothesis corresponded to no difference among groups. False discovery rate (FDR) was controlled by adjusting p-value using Benjamini-Hochberg algorithm. For a DEG set, hierarchical cluster analysis was performed using complete linkage and Euclidean distance as a measure of similarity. Gene-Enrichment and Functional Annotation analysis for significant probe list was performed using GO (www.geneontology.org) and KEGG (www.genome.jp/kegg). All data analyses and visualization of DEGs were conducted using R 3.1.2 (www.r-project.org). Statistical significance between the control group and experimental groups was determined using Student's t-test, Bonferroni correction, and FDR. In all cases, a p-value less than 0.05 was considered to be significant.
We used hierarchical clustering (Euclidean method, complete linkage) with respect to expression level (normalized value). Results showed that mRNA was differentially expressed between control and experimental groups (10%, 30% BMCF). Control group (Control_1–3), 10% group (Test10_1–3), and 30% group (Test30_1–3) had differential diagram each, hence proving that FIR affects gene expression in a dose-dependent manner in rat skeletal muscle (Fig. 1A). The results indicated that these genes showed similar expression trends in each group, as per the color key of blue and yellow. Based on microarray analysis, a total of 152 DEGs were identified in the control group v.s. 10% group, among which 114 genes were significantly up-regulated and 38 were down-regulated (FC > 1.5). In control group v.s. 30% group, 1473 DEGs were identified, among which 1,263 genes were significantly up-regulated and 210 were down-regulated (FC > 1.5) (Fig. 1B). Between the control and 30% groups, 218 genes were significantly regulated (FC > 1.5 and p < 0.05) (Fig. 1C).
Network analysis was performed on 22 significantly DEGs by microarray analysis. We showed the interaction relationship network of genes by STRING. The hub gene was identified through a centralized location in the interaction network. The hub genes identified in this study included Col3a1, Pdgfra, Spacr, and Actb (Fig. 2).
The 1,473 DEGs that were significantly stimulated in the 30% group were subjected to DAVID database and KEGG pathway mapper to investigate the functional interactions with FIR radiation in a dose-dependent manner. Based on KEGG map analysis, the significant differential expression levels in several signal pathways involved in focal adhesion, ECM-receptor interaction, proteoglycans in cancer and phagosome, etc. GO functions of the DEGs were determined according to different categories, including biological processes, molecular functions, and cellular components. In GO molecular function category of GO analysis, ECM binding (GO:0050840, 4.92971E-07) was significantly stimulated by DEGs, and single-organism process (GO:0044699, 1.20403E-43) and developmental process (GO:0032502, 3.05774E-36) were significantly stimulated on the GO analysis category. In cellular component category, extracellular region part (GO:0044421, 6.05992E-50) and vesicles (GO:0031982, 7.164E-44) were significantly enriched. Based on the GO and KEGG analysis, significantly stimulated pathways were listed according to cellular functions which involved in cell migration, ECM interaction, and PDGF-related pathway (Table 2).
Actin, integrin, and coronin, along with their subunits, were found up-regulated in microarray data. ECM and PDGF-mediated cell migration-related genes, such as Pdgfra, Pdgfrl, platelet factor 4 (Pf4), Pdgfd, phosphofructokinase platelet (Pfkp), thromboxane A synthase 1 platelet (Tbxas1), phospholipase A2 group IIA (Pla2g2a), actinin alpha 1 (Actn1), Actn4, actin gamma 1 (Actg1), Actb, Ecm1, Ecm2, collagen type 3 alpha 1 (Col3a1), Col6a1, laminin alpha 4 (Lama4), laminin beta 1 (Lamb1), laminin gamma 1 (Lamc1), Fn1, Itga2, Itga5, Itga11, integrin beta 5 (Itgb5), integrin beta-like 1 (Itgbl1), tropomyosin 3 (Tpm3), tropomyosin 4 (Tpm4), myosin heavy chain 3 (Myh3), and myosin heavy chain 9 (Myh9) were significantly increased. We re-verified the relevant mRNA expression via qRT-PCR (Fig. 3), and the results showed that a similar pattern to the FC value indicated in microarray analysis (Table 3).
Wound healing assay was performed to observe the migration of L6 cell line upon FIR radiation. Results showed that gap area of FIR groups at 24 h and 48 h in starvation medium was narrower than control group, suggesting the promotion of cell migration by FIR radiation (Fig. 4A). Fig. 4B showed the relative scratch gap area between control group (0%) and 10% group. And, the FIR radiation-induced wound healing effects were significantly increased at 24 h and 48 h (p < 0.01).
In this study, we demonstrated that FIR radiation affects the PDGF-mediated cell migration pathway in rats. In our previous study, we had established an experimental method to study the bio-active effects of FIR-radiated fabric on rats [37]. We considered rat skeletal muscle tissue and L6 cells for the effects of FIR radiation via microarray, qRT-PCR, and mRNA expression analysis of PDGF-cell migration-related genes, and verified their dose-dependence. FIR rays also up-regulated DEGs required for cell migration in actin-integrin signaling. Recent studies have shown that an improvement of various bio-activities, such as wound healing, deep skin heating, blood flow, and blood circulation by FIR radiation [12469101233]. All of these effects involve cell migration processes, and the wound healing process is particularly increased in this study (Table 2 and Fig. 4). However, the underlying mechanisms have not been clearly understood. FIR rays induce vibration of the water inside human bodies, thereby allowing its deeper penetration [2]. Near infrared radiation penetrates into the intracellular space and induces vibration in the cell membrane [40].
By microarray analysis, we observed that mRNA expression was stimulated by FIR radiation in a dose-dependent manner (Fig. 1). We also showed that ECM-receptor interaction, focal adhesion, and cell motility, in the cell migration-related pathway, were significantly stimulated (Table 2). Based on these results, we propose that FIR radiation can stimulate ECM and PDGF interaction. Recent studies have shown that the expression level of PDGF-receptor can be regulated by ECM signaling [4142]. PDGF-receptor-beta has been reported to mediate ECM-induced cell migration via tyrosine phosphorylation [31].
Interestingly, microarray results showed that ECM and PDGF-mediated cell migration-related genes were significantly increased and Pdgfra, Pdgfrl, Pf4, Pdgfd, Actn1, Actn4, Actg1, Actb, Ecm1, Ecm2, Col3a1, Col6a1, Col6a2, Col6a3, Lama4, Lamb1, Lamc1, Fn1, Itga2, Itga5, Itga11, Itgb5, Itgbl1, Myh3, and Myh9 were stimulated (Table 3). Since PDGF-related genes, a cell mobility target in FIR-mediated cell migration, were found to be stimulated, a significant induction of cell activity by FIR may be possible that may lead to an altered ECM, which could co-activate actin binding, eventually leading to the migration (Fig. 3) [43]. This may be explained by the vibration of cell membrane at high frequency upon FIR irradiation. Laminin, composed of alpha, beta, and gamma subunits, was stimulated by FIR radiation. Together with collagen 6, laminin forms the main structure of the basal layer and interacts with cell surface components and cellular receptors, such as integrin, to participate in cell attachment [44]. Collagen interacts with cell surface proteins, including integrins in ECM, and along with ECM components, col6a1, col6a2, col6a3, and col3a1 that determine tissue morphology and mechanical properties, are all up-regulated (Fig. 2) [45].
FIR rays stimulate actin subunit and actin-related genes, such as actinin, integrin, laminin, collagen, myosin, and fibronectin. Relative gene expression of Actb (core protein of cell composition) was found to be significantly increased in microarray analysis, although it was applied as a housekeeping gene. PDGF induces actin accumulation in cell membrane ruffles [46]. PDGF and VEGF receptors affect actin accumulation. F-actin accumulation would allow receptor activation to control cell migration [47]. Actin supports cell motility, -division, and -morphogenesis by intracellular protein interaction [33].
Up-regulated expression of Actb mRNA indicated that accumulated actin plays a pivotal role in wound healing process. Cell migration is reported to regulate wound healing by the assembly of actin for focal adhesion [48]. In this study, Actb expression did not considerably change in cellular experiments, but significantly increased in tissue experiment, with the use FIR-radiated fabric in a dose-dependent manner. Together, the results suggest that FIR radiation mediated wound healing by regulating cell components in skeletal muscle tissue.
We observed that wound healing was certainly promoted by FIR radiation over a span of 48 h (Fig. 4). Since FIR radiation efficacy had been proven in muscle tissue, we further checked whether it affects cells as well. Based on our results, we confirmed that FIR radiation affects not only tissues, but also cells. We have established experimental conditions for applying BMCF to cells in a subsequent study.
Therefore, our results propose that FIR radiation stimulates PDGF-mediated cell migration pathway via integrin-actin signaling. These mechanisms may prove useful for wound healingrelated treatment. In future, we plan to extend our study to measure the depolarization of membrane potential by a patch clamp method. Measurement of NO product is also required for verifying the expansion of blood vessels and increase in blood pressure, essential for microcirculation.
Figures and Tables
 | Fig. 1Microarray analysis of messenger RNA expression in bio-active materials-coated fabric and control rat.(A) Hierarchical clustering heatmap was classified as color based on differentially expressed genes between Test10 group (10% materials-coated fabric), Test30 groups (30% materials-coated fabric) and control group. Yellow and blue color represents higher and lower than z-score mean. (B) Up- and down-regulated genes count were indicated each group (fold change [FC] > 1.5 and 2). (C) Significant genes count was indicated each group (FC < 1.5 and < 2, p < 0.05).
|
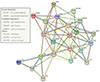 | Fig. 2Protein interaction network analysis.Protein interaction network was identified upon 22 differentially expressed genes via Search Tool for the Retrieval of Interacting Genes version 10.5 (STRING; www.string-db.org). Col6a1, collagen, type VI alpha 1; Col3a1, collagen, type 3, alpha 1; Nid1, nidogen 1; Bgn, biglycan; Mmp14, matrix metallopeptidase 14; Actb, actin beta; Mmp2, matrix metallopeptidase 2; Vim, vimentin; Cd34, cd34 molecule; Msn, moesin; Gsn, gelsolin; Lcp1, lymphocyte cytosolic protein 1; Bmp4, bone morphogenetic protein 4; Sparc, secreted protein acidic cysteinerich; Pdgfra, platelet-derived growth factor receptor alpha polypeptide; Igf1, insulin-like growth factor 1; Tgfb1, transforming growth factor beta 1; Plau, plasminogen activator urokinase; Lrp1, low density lipoprotein receptor-related protein 1; Csf1r, colony stimulating factor 1 receptor; Rarres2, retinoic acid receptor responder 2; Apoe, apolipoprotein E.
|
 | Fig. 3Expression of messenger RNA for platelet-derived growth factor (PDGF)-mediated cell migration-related genes.(A) PDGF D (Pdgfd), (B) PDGF receptor alpha (Pdgfra), (C) PDGF receptor-like (Pdgfrl), (D) extracellular matrix protein 1 (Ecm1), (E) extracellular matrix protein 2 (Ecm2), (F) matrix metallopeptidase 2 (Mmp2), (G) integrin alpha 5 (Itga5), and (H) actin beta (Actb). FIR, far-infrared. *Statistically significant vs. control group, p < 0.05; **statistically significant vs. control group, p < 0.01.
|
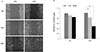 | Fig. 4Wound healing assay using L6 cells of 0% and 10% far-infrared (FIR) radiated fabric for 48 h.(A) Images of L6 cell for 0% and 10% FIR radiated fabric were captured upon 0, 24, and 48 h in wound healing assay. (B) Relative scratch gap area was based on 0 h in 0% materials-coated fabric group cell and which were calculated using Image J software. **Statistically significant vs. control group, p < 0.01.
|
Table 1
Primer used for quantitative real-time polymerase chain reaction

Pdgfrl, platelet-derived growth factor receptor-like; Pdgfd, platelet derived growth factor D; Pdgfra, platelet derived growth factor receptor alpha polypeptide; Ecm1, extracellular matrix protein 1; Ecm2, extracellular matrix protein 2; Fn1, fibronectin 1; Actb, actin beta; Mmp14, matrix metallopeptidase 14; Mmp2, matrix metallopeptidase 2; RhoB, ras homolog family member B; RhoJ, ras homolog family member J; Itga5, integrin alpha 5; Itga11, integrin alpha 11.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (No. NRF-2017R1A2B4002052). This research was supported by the Chung-Ang University Research Scholarship Grants in 2018.
Notes
References
1. Yu SY, Chiu JH, Yang SD, Hsu YC, Lui WY, Wu CW. Biological effect of far-infrared therapy on increasing skin microcirculation in rats. Photodermatol Photoimmunol Photomed. 2006; 22:78–86.


2. Beever R. Far-infrared saunas for treatment of cardiovascular risk factors: summary of published evidence. Can Fam Physician. 2009; 55:691–696.


3. Adamskaya N, Dungel P, Mittermayr R, Hartinger J, Feichtinger G, Wassermann K, Redl H, van Griensven M. Light therapy by blue LED improves wound healing in an excision model in rats. Injury. 2011; 42:917–921.

4. Vatansever F, Hamblin MR. Far infrared radiation (FIR): its biological effects and medical applications. Photonics Lasers Med. 2012; 4:255–266.



5. Tsai SR, Hamblin MR. Biological effects and medical applications of infrared radiation. J Photochem Photobiol B. 2017; 170:197–207.



6. Inoué S, Kabaya M. Biological activities caused by far-infrared radiation. Int J Biometeorol. 1989; 33:145–150.


7. Rau CS, Yang JC, Jeng SF, Chen YC, Lin CJ, Wu CJ, Lu TH, Hsieh CH. Far-infrared radiation promotes angiogenesis in human microvascular endothelial cells via extracellular signal-regulated kinase activation. Photochem Photobiol. 2011; 87:441–446.


8. Tran TH, Mai HN, Shin EJ, Nam Y, Nguyen BT, Lee YJ, Jeong JH, Tran HY, Cho EH, Nah SY, Lei XG, Nabeshima T, Kim NH, Kim HC. Repeated exposure to far infrared ray attenuates acute restraint stress in mice via inhibition of JAK2/STAT3 signaling pathway by induction of glutathione peroxidase-1. Neurochem Int. 2016; 94:9–22.


9. Huang PH, Chen JW, Lin CP, Chen YH, Wang CH, Leu HB, Lin SJ. Far infra-red therapy promotes ischemia-induced angiogenesis in diabetic mice and restores high glucose-suppressed endothelial progenitor cell functions. Cardiovasc Diabetol. 2012; 11:99.



10. Yang CS, Yeh CH, Tung CL, Chen MY, Jiang CH, Yeh ML. Impact of far-infrared ray exposure on the mechanical properties of unwounded skin of rats. Exp Biol Med (Maywood). 2010; 235:952–956.


11. Shui S, Wang X, Chiang JY, Zheng L. Far-infrared therapy for cardiovascular, autoimmune, and other chronic health problems: a systematic review. Exp Biol Med (Maywood). 2015; 240:1257–1265.


12. Hsu YH, Chen YC, Chen TH, Sue YM, Cheng TH, Chen JR, Chen CH. Far-infrared therapy induces the nuclear translocation of PLZF which inhibits VEGF-induced proliferation in human umbilical vein endothelial cells. PLoS One. 2012; 7:e30674.

13. Noponen PVA, Häkkinen K, Mero AA. Effects of far infrared heat on recovery in power athletes. J Athl Enhancement. 2015; 4:DOI: 10.4172/2324-9080.1000202.

14. Mai HN, Sharma N, Shin EJ, Nguyen BT, Nguyen PT, Jeong JH, Jang CG, Cho EH, Nah SY, Kim NH, Nabeshima T, Kim HC. Exposure to far-infrared rays attenuates methamphetamine-induced recognition memory impairment via modulation of the muscarinic M1 receptor, Nrf2, and PKC. Neurochem Int. 2018; 116:63–76.


15. Chang HY, Shih MH, Huang HC, Tsai SR, Juan HF, Lee SC. Middle infrared radiation induces G2/M cell cycle arrest in A549 lung cancer cells. PLoS One. 2013; 8:e54117.

16. Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003; 302:1704–1709.


17. Sheetz MP, Felsenfeld DP, Galbraith CG. Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol. 1998; 8:51–54.


18. Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002; 2:91–100.


19. Krawczyk C, Oliveira-dos-Santos A, Sasaki T, Griffiths E, Ohashi PS, Snapper S, Alt F, Penninger JM. Vav1 controls integrin clustering and MHC/peptide-specific cell adhesion to antigen-presenting cells. Immunity. 2002; 16:331–343.


20. Blazevic T, Schwaiberger AV, Schreiner CE, Schachner D, Schaible AM, Grojer CS, Atanasov AG, Werz O, Dirsch VM, Heiss EH. 12/15-lipoxygenase contributes to platelet-derived growth factor-induced activation of signal transducer and activator of transcription 3. J Biol Chem. 2013; 288:35592–35603.
21. Alvarez RH, Kantarjian HM, Cortes JE. Biology of platelet-derived growth factor and its involvement in disease. Mayo Clin Proc. 2006; 81:1241–1257.


22. Yu J, Moon A, Kim HR. Both platelet-derived growth factor receptor (PDGFR)-alpha and PDGFR-beta promote murine fibroblast cell migration. Biochem Biophys Res Commun. 2001; 282:697–700.

23. Robbins JR, McGuire PG, Wehrle-Haller B, Rogers SL. Diminished matrix metalloproteinase 2 (MMP-2) in ectomesenchyme-derived tissues of the Patch mutant mouse: regulation of MMP-2 by PDGF and effects on mesenchymal cell migration. Dev Biol. 1999; 212:255–263.


24. Xu J, Clark RA. Extracellular matrix alters PDGF regulation of fibroblast integrins. J Cell Biol. 1996; 132:239–249.



25. Zhang F, Hao F, An D, Zeng L, Wang Y, Xu X, Cui MZ. The matricellular protein Cyr61 is a key mediator of platelet-derived growth factor-induced cell migration. J Biol Chem. 2015; 290:8232–8242.



26. Li J, Kim YN, Bertics PJ. Platelet-derived growth factor-stimulated migration of murine fibroblasts is associated with epidermal growth factor receptor expression and tyrosine phosphorylation. J Biol Chem. 2000; 275:2951–2958.


27. Raines EW, Lane TF, Iruela-Arispe ML, Ross R, Sage EH. The extracellular glycoprotein SPARC interacts with platelet-derived growth factor (PDGF)-AB and -BB and inhibits the binding of PDGF to its receptors. Proc Natl Acad Sci U S A. 1992; 89:1281–1285.



28. Yuan L, Santi M, Rushing EJ, Cornelison R, MacDonald TJ. ERK activation of p21 activated kinase-1 (Pak1) is critical for medulloblastoma cell migration. Clin Exp Metastasis. 2010; 27:481–491.



29. MacDonald TJ, Brown KM, LaFleur B, Peterson K, Lawlor C, Chen Y, Packer RJ, Cogen P, Stephan DA. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet. 2001; 29:143–152.


30. Borkham-Kamphorst E, van Roeyen CR, Ostendorf T, Floege J, Gressner AM, Weiskirchen R. Pro-fibrogenic potential of PDGF-D in liver fibrosis. J Hepatol. 2007; 46:1064–1074.


31. Veevers-Lowe J, Ball SG, Shuttleworth A, Kielty CM. Mesenchymal stem cell migration is regulated by fibronectin through α5β1-integrin-mediated activation of PDGFR-β and potentiation of growth factor signals. J Cell Sci. 2011; 124:1288–1300.



32. Schlaepfer DD, Hanks SK, Hunter T, van der Geer P. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature. 1994; 372:786–791.


33. Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008; 9:344–356.

34. Vicente-Manzanares M, Choi CK, Horwitz AR. Integrins in cell migration--the actin connection. J Cell Sci. 2009; 122:199–206.

36. Hsu YH, Lin YF, Chen CH, Chiu YJ, Chiu HW. Far infrared promotes wound healing through activation of Notch1 signaling. J Mol Med (Berl). 2017; 95:1203–1213.


37. Lee D, Kim YW, Kim JH, Yang M, Bae H, Lim I, Bang H, Go KC, Yang GW, Rho YH, Park HS, Park EH, Ko JH. Improvement characteristics of bio-active materials coated fabric on rat muscular mitochondria. Korean J Physiol Pharmacol. 2015; 19:283–289.



38. Lee SG, Lee CG, Wu HM, Oh CS, Chung SW, Kim SG. A load of mice to hypergravity causes AMPKα repression with liver injury, which is overcome by preconditioning loads via Nrf2. Sci Rep. 2015; 5:15643.



39. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–D452.

40. Henderson TA, Morries LD. Near-infrared photonic energy penetration: can infrared phototherapy effectively reach the human brain? Neuropsychiatr Dis Treat. 2015; 11:2191–2208.



41. Zhu J, Clark RAF. Fibronectin at select sites binds multiple growth factors and enhances their activity: expansion of the collaborative ECM-GF paradigm. J Invest Dermatol. 2014; 134:895–901.


42. Laurent M, Martinerie C, Thibout H, Hoffman MP, Verrecchia F, Le Bouc Y, Mauviel A, Kleinman HK. NOVH increases MMP3 expression and cell migration in glioblastoma cells via a PDGFR-alphadependent mechanism. FASEB J. 2003; 17:1919–1921.

43. Hardee JP, Puppa MJ, Fix DK, Gao S, Hetzler KL, Bateman TA, Carson JA. The effect of radiation dose on mouse skeletal muscle remodeling. Radiol Oncol. 2014; 48:247–256.



44. Miner JH, Patton BL, Lentz SI, Gilbert DJ, Snider WD, Jenkins NA, Copeland NG, Sanes JR. The laminin alpha chains: expression, developmental transitions, and chromosomal locations of alpha1-5, identification of heterotrimeric laminins 8-11, and cloning of a novel alpha3 isoform. J Cell Biol. 1997; 137:685–701.
45. Boraschi-Diaz I, Wang J, Mort JS, Komarova SV. Collagen type I as a ligand for receptor-mediated signaling. Front Phys. 2017; 5:DOI: 10.3389/fphy.2017.00012.

46. Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992; 70:389–399.






 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download