1. Aly AH, Debbab A, Proksch P. Fungal Divers. 2011; 50:3–19.
2. Wiemann P, Keller NP. J Ind Microbiol Biotechnol. 2014; 41:301–313.
3. Matsuda Y, Abe I. Nat Prod Rep. 2016; 33:26–53.
4. Schueffler A, Anke T. Nat Prod Rep. 2014; 31:1425–1448.
5. Chexal KK, Springer JP, Clardy J, Cole RJ, Kirksey JW, Dorner JW, Cutler HG, Strawter BJ. J Am Chem Soc. 1976; 98:6748–6750.
6. Geris R, Simpson T. Nat Prod Rep. 2009; 26:1063–1094.
7. Matsuda Y, Awakawa T, Wakimoto T, Abe I. J Am Chem Soc. 2013; 135:10962–10965.
8. Matsuda Y, Iwabuchi T, Fujimoto T, Awakawa T, Nakashima Y, Mori T, Zhang H, Hayashi F, Abe I. J Am Chem Soc. 2016; 138:12671–12677.
9. Matsuda Y, Awakawa T, Mori T, Abe I. Curr Opin Chem Biol. 2016; 31:1–7.
10. Mosmann T. J Immunol Methods. 1983; 65:55–63.
11. Ulukaya E, Ozdikicioglu F, Oral AY, Demirci M. Toxicol In Vitro. 2008; 22:232–239.
12. Johansson M, Karlsson L, Wennergren M, Jansson T, Powell TL. J Clin Endocrinol Metab. 2003; 88:2831–2837.
13. Oh KB, Kim SH, Lee J, Cho WJ, Lee T, Kim S. J Med Chem. 2004; 47:2418–2421.
14. Lee HS, Lee TH, Yang SH, Shin HJ, Shin J, Oh KB. Bioorg Med Chem Lett. 2007; 17:2483–2486.
15. Oh KB, Lee JH, Chung SC, Shin J, Shin HJ, Kim HK, Lee HS. Bioorg Med Chem Lett. 2008; 18:104–108.
16. dos Santos RM, Rodrigues-Fo E. Z Naturforsch C. 2003; 58:663–669.
17. Fill TP, Pereira GK, Geris dos Santos RM, Rodrigues-Fo E. Z Naturforsch. 2007; 62b:1035–1044.
18. Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, Chiang YM, Oakley BR, Wang CC. J Am Chem Soc. 2012; 134:4709–4720.
19. Arunpanichlert J, Rukachaisirikul V, Phongpaichit S, Supaphon O, Sakayaroj J. Tetrahedron. 2015; 71:882–888.
20. Hayashi H, Mukaihara M, Murao S, Arai M, Lee AY, Clardy J. Biosci Biotech Biochem. 1994; 58:334–338.
21. Fayos J, Lokensgard D, Clardy J, Cole RJ, Kirksey JW. J Am Chem Soc. 1974; 96:6785–6787.
22. Simpson TJ, Stenzel DJ, Bartlett AJ, O'Brien E, Holker JSE. J Chem Soc Perkin Trans 1. 1982; 2687–2692.
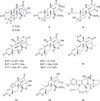
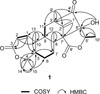
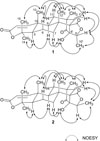
 -26.9 (c 0.20, CHCl3); UV (MeOH) λmax (log ε) 211 (3.49) nm; 1H and 13C NMR: see Table 1; IR (ZnSe) νmax 3412, 2973, 2938, 1758, 1694 cm−1; HRFABMS, m/z 429.2275 [M+H]+ (calcd for C25H33O6, 429.2277).
-26.9 (c 0.20, CHCl3); UV (MeOH) λmax (log ε) 211 (3.49) nm; 1H and 13C NMR: see Table 1; IR (ZnSe) νmax 3412, 2973, 2938, 1758, 1694 cm−1; HRFABMS, m/z 429.2275 [M+H]+ (calcd for C25H33O6, 429.2277). -50.9 (c 0.20, CHCl3); UV (MeOH) λmax (log ε) 212 (3.83) nm; 1H and 13C NMR: see Table 1; IR (ZnSe) νmax 3120, 2973, 2931, 1755, 1687 cm−1; HRFABMS, m/z 429.2279 [M+H]+ (calcd for C25H33O6, 429.2277).
-50.9 (c 0.20, CHCl3); UV (MeOH) λmax (log ε) 212 (3.83) nm; 1H and 13C NMR: see Table 1; IR (ZnSe) νmax 3120, 2973, 2931, 1755, 1687 cm−1; HRFABMS, m/z 429.2279 [M+H]+ (calcd for C25H33O6, 429.2277).



 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download