Abstract
Apolipoprotein E is a plasma protein that has an important role in transport and metabolism of lipids in serum as well as central nervous system. Among the 3 common alleles, the ε2 allele has the most stable structure followed by ε3 and ε4 in order. There is evidence for a deleterious role of ε4 allele by atherosclerosis and amyloid beta accumulation in brain and body. The presence and gene dose of ε4 allele are risk factors for late-onset Alzheimer's disease. Apolipoprotein E ε4 may have a role in the pathology of amyloid beta and tau and it has a strong relationship with the early onset of late-onset Alzheimer's disease. However, early-onset Alzheimer's disease has a weaker relationship with ε4 allele of apolipoprotein E.
The human apolipoprotein E (apoE, protein; APOE, gene) is single chain protein, classified as a lipoprotein, with 299 amino acids. ApoE has 2 domains i.e., the amino-terminal domain that has low-density lipoprotein receptor binding region and a carboxy-terminal domain that has lipid-binding region.1 APOE gene, located on chromosome 19q13, has several single-nucleotide polymorphisms.2 The 3 common type polymorphisms are ε2, ε3, ε4; whereas rare type polymorphisms include ε1, ε5, ε7. The common types constitute 3 homozygous (ε2/ε2, ε3/ε3, ε4/ε4) and 3 heterozygous (ε2/ε3, ε2/ε4, ε3/ε4) diplotypes, which induces either exchange of 1 or 2 amino acid(s)34 or glycosylation of 1 amino acid.5 The ε2, ε3, and ε4 alleles differ by single amino acid substitutions at residues 112 and 158 of the protein. The amino acids sequence of these residues is cysteine-cysteine (ε2), cysteine-arginine (ε3), and arginine-arginine (ε4).6 The ε3 allele also is the most common allele in all population and neutral or protective to cells and organs.
ApoE is an important plasma protein found in plasma lipids such as very low-density lipoproteins, chylomicron, and a subclass of high-density lipoprotein. It is essential for the catabolism of triglyceride-rich lipoprotein constituents, transportation of cholesterol and other lipids, and cellular repair.78 Foods with high cholesterol and triglyceride induce its expression in various animals.9 The liver is the main organ producing apoE in human, producing >75% of total apoE. Brain, spleen, lung, kidney, ovary, testis, peripheral nerves and muscle also produce apoE.10 Cholesterol is associated with myelin production and essential component of the brain cell membrane. It contributes to brain development, neuronal maintenance, and repair, as well as maintaining the synaptic plasticity of neuron cells.11 Astrocytes and microglia, vascular smooth muscle cells, and choroid plexus are sources of apoE in the human brain. Neurons can produce apoE, especially under stressful conditions.712 Increased apoE can modulate lipid metabolism in the compromised nervous system.
APOE polymorphism was identified in 1993 in relationship to the onset and clinical feature of Alzheimer's disease. It has since become an important factor in the understanding of pathophysiology of Alzheimer's disease, immunoregulation, and cognition in other dementias.13
Whether APOE has a protective or harmful role in the brain is under debate.1415 In an epidemiological study, the terms are relative.161718 When the APOE ε4 allele frequency is higher in patients with Alzheimer's disease than cognitively normal person, APOE ε4 is considered harmful. On the other hand, when the frequency of APOE ε2 allele in Alzheimer's disease patients is less than that of cognitively normal person, APOE ε2 allele may be protective.19 However, these terms have a different meaning in laboratory studies. When cells with APOE ε2 allele survive longer than cells with other haplotypes in a toxic environment, then APOE ε2 is protective. APOE ε4 is considered toxic to the nervous system and vascular endothelial cells, as compared to the other isoforms. The biological efficacy of APOE ε3 is between APOE ε2 and APOE ε4, hence, APOE ε3 is considered neutral in terms of risk for Alzheimer's disease. 1420
The mechanisms for the harmful effect of APOE ε4 are as follows. First, "domain interaction" theory explains the negative role of the APOE ε4.21 The domain interaction occurs between Arg-61 of the amino domain and Glu-255 of the carboxy-domain. This single amino acid interchange of the APOE ε4 causes a structural change such that APOE ε4 becomes more compact than APOE ε3 or APOE ε2.22 This mediates the adverse effects of APOE ε4 (Fig. 1).23 Second, affinity of APOE ε4 for very low-density lipoproteins and low-density lipoprotein could explain brain damage by APOE ε4.24 Third, a recent study confirmed that proteolytically cleaved APOE ε4 is a major factor in Alzheimer's disease. An amino-terminal fragment of APOE ε4 is identified in neurofibrillary tangles using antibody, suggestive of neurotoxic effect of the amino terminal.2526 Finally, the carboxy-domain fragments of APOE ε4 are neurotoxic and cause mitochondrial dysfunction and formation of neurofibrillary tangles in transgenic mice.27
Alzheimer's disease is the most common cause of dementia in the elderly.28 With an increment of life expectancy in developed countries, the incidence and prevalence of Alzheimer's disease are significantly rising. The prevalence of Alzheimer's disease is increasing roughly at 2-fold rate per 5 years in patients above 65 years of age, reaching >30% at age 85. Alzheimer's disease is considered to have heterogeneous genetic causes. It is divided into early-onset Alzheimer's disease and late-onset Alzheimer's disease by the age 65. Interestingly, Alzheimer's disease with the strong genetic background, usually autosomal dominant, has a relatively early onset of around 50 years. Moreover, research with genome-wide association study reveals that APOE ε4 carriers have a 33-fold higher risk of Alzheimer's disease than APOE ε3/3 carriers.2930 Loss of short term memory is the earliest clinical feature followed by loss of other cognitive features such as visuospatial function, language function, and frontal executive function. The primary pathology of Alzheimer's disease is an abnormal aggregation of amyloid beta that is a produced from amyloid precursor protein31 in the extracellular space and tau protein in the neuronal cell.32 Accumulation of amyloid beta causes senile plaque and accumulation of abnormal tau protein causes neurofibrillary tangle. The majoir component of neurofibrillary tangles is hyperphosphorylated tau, a form of paired helical filament.333435 APOE ε4 is precisely correlated with cerebrospinal fluid amyloid beta levels in the preclinical stage of Alzheimer's disease, which is less prominent in full-blown dementia.36 APOE ε4 may also mediate the development of dementia through tau phosphorylation, destruction of cytoskeleton, and mitochondrial dysfunction. 373839 Experiments with cellular models, animal models, and patient biomarkers suggest that amyloid beta induces tau pathology. However, the relationship between amyloid beta and tau protein and their respective role(s) in Alzheimer's disease remains unclear.4041 Perivascular accumulation of amyloid beta also leads to other pathologies such as cerebral amyloid angiopathy.42 Longitudinal neuroimaging and pathological studies show that pathological changes of Alzheimer's disease begin decades before the clinical onset.43444546 Excess aggregation of amyloid beta is a major shift in early stage Alzheimer's disease. Amyloid beta 40 and 42 are important components among its subtypes. Amyloid beta 40 is more prevalent and less toxic than Amyloid beta 42.47 Amyloid beta associated senile plaques and hyperphosphorylated tau associated neurofibrillary tangles are possibly associated with APOE ε4.48495051 These pathological changes result in loss of dendritic spines and decrement of synaptic density, finally, neuronal cells' death.5253 While the former study supports a harmful role of APOE ε4,373839 later study suggests that multiple factors modulate the effect of APOE ε4 in the development of Alzheimer's disease.5051
Three recent studies explained the discrepancy between the amount of amyloid beta and cognitive dysfunction. A study of gene expression in the cerebral cortex of APOE ε4 carriers and late-onset Alzheimer's disease indicates several regulatory mediators including APBA2, FYN, RNF219, and SV2A of which, those involved in amyloid beta precursor protein metabolism are likely to be associated with pathologic changes in late-onset Alzheimer's disease.54 The longitudinal study, Alzheimer Disease Neuroimaging Initiative (http://www.adni-info.org) likewise shows that APOE ε4 participates in the pathology of preclinical Alzheimer's disease via amyloid beta. They also found a significant relationship between cerebrospinal fluid amyloid beta and cerebrospinal fluid clusterin as well as cerebrospinal fluid amyloid beta and cerebrospinal fluid phosphorylated tau on entorhinal cortex atrophy rate. Thus, phosphorylated tau protein and clusterin, a chaperone glycoprotein, mediate neurodegeneration. 55
Thirdly, hippocampal oscillation of theta and gamma rhythms are possibly associated with cognition. Animal models indicate that hippocampal and cortical network undergo reorganization in Alzheimer's disease. Altered oscillation of theta and gamma rhythm develops first followed by increased amyloid burden, and finally loss of gamma-amino-butyricacidergic neurons. Moreover, high levels of amyloid beta in hippocampus cause seizure activity without serious neuronal loss.5657
Late-onset Alzheimer's disease is multifactorial, including genetic and environmental factors with negative impact on endocytic function, lipoprotein signaling as well as synaptic regulation.60 Recently, chronic inflammation causing focal accumulation of mitochondria suggested as a triggering factor for late-onset Alzheimer's disease.61 Usually, late-onset Alzheimer's disease develops after the age of 65 years, and 60 years is proposed as the more appropriate cut-off age of the illness. 2862 Amyloid plaque and neurofibrillary tangle are the main pathological findings of late-onset Alzheimer's disease. APOE ε4 occurs in up to 80% of late-onset Alzheimer's disease patients and is considered a risk factor for this dementing illness. 131662 The following study shows that apoE strongly binds amyloid beta and APOE ε4 is the common haplotype in lateonset Alzheimer's disease.49 Moreover, carriers of APOE ε4 showed up to 15 years earlier disease onset and increased incidence of neuropsychiatric symptoms.6364 Intriguingly, two African populations with high frequencies of APOE ε4 show no such strong relationship.3065 In late-onset Alzheimer's disease, the function of key amyloid beta processing enzymes is normal. 60 However, the amount of amyloid plaque in the brain increases not because of increased amyloid beta production but because of impaired clearance.66 APOE ε4 is responsible for reduced amyloid clearance in the diseased brain.67 The presence of APOE ε4 also related with more rapid progression and poor response to cholinergic therapy in many ethnic groups.13 However, results are unequivocal in only Caucasian populations; adequate evidence in other ethnic groups such as African American and Hispanic populations is still needed. Relatively smaller sample size, allele frequency variation among ethnicities, and lifestyle issues could explain the discrepancy.18686970 Overall, APOE polymorphism is not a useful diagnostic biomarker or prognostic factor for late-onset Alzheimer's disease, as compared to amyloid beta 42 and tau in cerebrospinal fluid. 71 However, it still may be used as a predictor of increased neuropsychiatric symptoms and decreased response to pharmacological therapy.131564
Early-onset Alzheimer's disease develops before 65 years old, and it is rare disease composing <1% of Alzheimer's disease cases.62 Alzheimer's disease was first reported by Dr. Alois Alzheimer in the early 20th century as a case of early-onset disease.72 Since Corder et al.13 reported APOE ε4 as a risk factor for late-onset Alzheimer's disease, much effort is made to clarify the relationship between early-onset Alzheimer's disease and APOE ε4. However, the results are inconclusive. Instead, other genes affecting amyloid precursor protein processing are highlighted and evaluated for a possible relation with early-onset Alzheimer's disease.62 These include amyloid precursor protein, presenilin-1, and presenilin-2, which are associated with early-onset autosomal dominant Alzheimer's disease. Mutations in amyloid precursor protein gene are related to the conversion of amyloid precursor protein to a better substrate of beta-secretase. Amyloid beta derived from mutant amyloid precursor protein is more easily aggregated than that from wild type. Patient with presenilin mutation usually develops Alzheimer's disease between 30 and 50 years of age. Presenilin mutations were initially thought to increase gammasecretase activity. However, recent studies reveal that these mutations decrease gamma-secretase activity but increase the ratio of amyloid-beta42/amyloid-beta40, which supports the loss of function hypothesis.73
In contrast to the 2 enzymes, alpha-secretase reduces amyloid beta production in the brain. Increased brain APOE ε4 has an association with enhanced beta--secretase activity and subsequently increased amyloid beta production.54 Endosome dysfunction is now considered to have a major role in the production of large amount of amyloid beta in sporadic Alzheimer's disease. A postmortem study shows that enlarged endosomes facilitate a higher chance of amyloid cleavage by beta and gamma-secretase before the development of clinical dementia in APOE carriers.74 However, endosomal abnormalities are absent in early-onset familial Alzheimer's disease.75 This result suggests differential mechanisms between early-onset and late-onset Alzheimer's disease. Overall, APOE polymorphism appears to have a limited role in early-onset Alzheimer's disease.
APOE polymorphism in various ethnic groups is based on the specific disease-status of the group. APOE status follows Mendelian inheritance, with regional as well as ethnic difference. In a haplotype analysis study, APOE ε4 was suggested as the ancestral allele in humans.76 According to this theory, APOE ε3 and APOE ε2 evolved from APOE ε4, but APOE ε4 remained after this evolution. Interestingly, apoE amino acid sequence of chimpanzee, genetically closest to humans, is monomorphic, similar to the human APOE ε3.77 Reduced frequency of APOE ε4 is a major factor for increased human lifespan with a risk reduction of Alzheimer's disease and cardiovascular disease.30 As the amount of dietary fat and cholesterol increased during the ancient history of mankind, APOE ε3, which can reduce increased cholesterol level with APOE ε4, may evolve.78 Allele frequency of APOE ε4 in human population is uneven, with high frequencies of APOE ε4 in the equatorial area and high latitudes areas.79
In a Korean study of patients with Alzheimer's disease, the most common APOE allele is APOE ε3 (71.3%), followed by APOE ε4 (21.3%), APOE ε2 (7.5%).18 A population study with normal elderly Korean showed that the most common APOE allele is APOE ε3 (86.9%), followed by APOE ε4 (6.6%), APOE ε2 (6.5%).80
In general, Caucasians and Africans have higher frequencies of APOE ε4 than Asians. APOE ε3 is most commonly found in the majority of populations with a range of 8.5 to 98 percent, followed by ε4 (0 to 50%), and ε2 (0 to 37.5%).1879 These variations of APOE polymorphism among areas in the world and ethnicities could affect the results of clinical studies and drug efficacies.
APOE interacts with environmental and genetic factors in the onset and progression of neurodegenerative diseases as well as cognitive decline. Genotyping APOE polymorphism is a traditional method for evaluation of dementia and neurodegenerative diseases. Evidence strongly suggests APOE ε4 as a risk factor for late-onset Alzheimer's disease, but the low predictive value prevents it from usage in diagnosis and prognosis. There is less evidence of APOE genotype as a risk factor for early-onset Alzheimer's disease. Although APOE genotyping itself does not precisely predict the development of a particular disease, it can be included in the fully integrated evaluation of neurological diseases. Preventing the toxic effect of APOE ε4 may be a method of prevention and treatment of dementing illnesses.
Figures and Tables
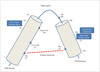 | Fig. 1Schematic diagram of human apolipoprotein E structure and main functional areas. The NH2 terminal domain and COOH terminal domain is connected by a flexible hinge region. There is low-density lipoprotein receptor binding region in NH2 terminal domain while Major lipid binding region and Amyloid beta interaction region are in COOH terminal domain. In apolipoprotein E ε4, domain reaction occurs between Arginine at residue 61 and Glutamate at residue 255 which results in the compact shape of the molecule. LDL: low-density lipoprotein. |
Acknowledgements
I wish to express my gratitude to Dr. Youn, Young Chul in Chung-Ang University for his good advice in revising this article.
References
1. Corbo RM, Prévost M, Raussens V, Gambina G, Moretto G, Scacchi R. Structural and phylogenetic approaches to assess the significance of human Apolipoprotein E variation. Mol Genet Metab. 2006; 89:261–269.

2. Nickerson DA, Taylor SL, Fullerton SM, Weiss KM, Clark AG, Stengård JH, et al. Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res. 2000; 10:1532–1545.

3. Park CH, Lee ST, Ki CS, Kim JW. Discrepancy in genotyping of apolipoprotein E between allele-specific PCR and fluorescence resonance energy transfer or sequencing. Korean J Lab Med. 2010; 30:325–328.

4. Scacchi R, Gambina G, Ferrari G, Corbo RM. Screening of two mutations at exon 3 of the apolipoprotein E gene (sites 28 and 42) in a sample of patients with sporadic late-onset Alzheimer's disease. Neurobiol Aging. 2003; 24:339–343.

5. Wernette-Hammond ME, Lauer SJ, Corsini A, Walker D, Taylor JM, Rall SC Jr. Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. J Biol Chem. 1989; 264:9094–9101.

6. Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994; 45:249–302.

7. Mahley RW, Huang Y. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 2012; 76:871–885.

8. Mahley RW, Huang Y. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr Opin Lipidol. 1999; 10:207–217.

9. Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J Lipid Res. 2009; 50:Suppl. S183–S188.

10. Zhang H, Wu LM, Wu J. Cross-talk between apolipoprotein E and cytokines. Mediators Inflamm. 2011; 2011:949072.

11. Kim J, Yoon H, Basak J, Kim J. Apolipoprotein E in synaptic plasticity and Alzheimer's disease: potential cellular and molecular mechanisms. Mol Cells. 2014; 37:767–776.

12. Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, Huang Y. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci. 2006; 26:4985–4994.

13. Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993; 261:921–923.

14. Rebeck GW, Kindy M, LaDu MJ. Apolipoprotein E and Alzheimer's disease: the protective effects of ApoE2 and E3. J Alzheimers Dis. 2002; 4:145–154.

15. Villeneuve S, Brisson D, Marchant NL, Gaudet D. The potential applications of Apolipoprotein E in personalized medicine. Front Aging Neurosci. 2014; 6:154.

16. Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997; 278:1349–1356.

17. van Duijn CM, de Knijff P, Cruts M, Wehnert A, Havekes LM, Hofman A, et al. Apolipoprotein E4 allele in a population-based study of early-onset Alzheimer's disease. Nat Genet. 1994; 7:74–78.

18. Kwon OD, Khaleeq A, Chan W, Pavlik VN, Doody RS. Apolipoprotein E polymorphism and age at onset of Alzheimer's disease in a quadriethnic sample. Dement Geriatr Cogn Disord. 2010; 30:486–491.

19. Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994; 7:180–184.

20. Wu YN, Zhang JW, Zhang ZX, Qu QM, Chen D. [An association analysis of apolipoprotein E genotypes with Alzheimer's disease in Chinese population]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2001; 23:450–454.

21. Mahley RW, Huang Y. Alzheimer disease: multiple causes, multiple effects of apolipoprotein E4, and multiple therapeutic approaches. Ann Neurol. 2009; 65:623–625.

22. Raffai RL, Dong LM, Farese RV Jr, Weisgraber KH. Introduction of human apolipoprotein E4 "domain interaction" into mouse apolipoprotein E. Proc Natl Acad Sci U S A. 2001; 98:11587–11591.

23. Ye S, Huang Y, Müllendorff K, Dong L, Giedt G, Meng EC, et al. Apolipoprotein (apo) E4 enhances amyloid beta peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc Natl Acad Sci U S A. 2005; 102:18700–18705.

24. Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006; 31:445–454.

25. Rohn TT. Proteolytic cleavage of apolipoprotein E4 as the keystone for the heightened risk associated with Alzheimer's disease. Int J Mol Sci. 2013; 14:14908–14922.

26. Rohn TT, Catlin LW, Coonse KG, Habig JW. Identification of an amino-terminal fragment of apolipoprotein E4 that localizes to neurofibrillary tangles of the Alzheimer's disease brain. Brain Res. 2012; 1475:106–115.

27. Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci U S A. 2005; 102:18694–18699.

29. Tanzi RE. The genetics of Alzheimer disease. Cold Spring Harb Perspect Med. 2012; 2(10):pii: a006296. DOI: 10.1101/cshperspect.a006296.

30. Raichlen DA, Alexander GE. Exercise, APOE genotype, and the evolution of the human lifespan. Trends Neurosci. 2014; 37:247–255.

31. Golde TE, Eckman CB, Younkin SG. Biochemical detection of Abeta isoforms: implications for pathogenesis, diagnosis, and treatment of Alzheimer's disease. Biochim Biophys Acta. 2000; 1502:172–187.

32. Lee VM, Trojanowski JQ. The disordered neuronal cytoskeleton in Alzheimer's disease. Curr Opin Neurobiol. 1992; 2:653–656.

33. Golde TE, Petrucelli L, Lewis J. Targeting Abeta and tau in Alzheimer's disease, an early interim report. Exp Neurol. 2010; 223:252–266.
34. Selkoe DJ. Clearing the brain's amyloid cobwebs. Neuron. 2001; 32:177–180.
36. Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Shaw LM, Trojanowski JQ, et al. Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Ann Neurol. 2010; 67:308–316.

37. Harris FM, Brecht WJ, Xu Q, Mahley RW, Huang Y. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: modulation by zinc. J Biol Chem. 2004; 279:44795–44801.

38. Tesseur I, Van Dorpe J, Spittaels K, Van den Haute C, Moechars D, Van Leuven F. Expression of human apolipoprotein E4 in neurons causes hyperphosphorylation of protein tau in the brains of transgenic mice. Am J Pathol. 2000; 156:951–964.

39. Agosta F, Vossel KA, Miller BL, Migliaccio R, Bonasera SJ, Filippi M, et al. Apolipoprotein E epsilon4 is associated with disease-specific effects on brain atrophy in Alzheimer's disease and frontotemporal dementia. Proc Natl Acad Sci U S A. 2009; 106:2018–2022.

40. Stancu IC, Vasconcelos B, Terwel D, Dewachter I. Models of β-amyloid induced Tau-pathology: the long and "folded" road to understand the mechanism. Mol Neurodegener. 2014; 9:51.

41. Inbar D, Belinson H, Rosenman H, Michaelson DM. Possible role of tau in mediating pathological effects of apoE4 in vivo prior to and following activation of the amyloid cascade. Neurodegener Dis. 2010; 7:16–23.

42. Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002; 297:353–356.

43. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991; 82:239–259.

44. Frisoni GB, Ganzola R, Canu E, Rüb U, Pizzini FB, Alessandrini F, et al. Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 Tesla. Brain. 2008; 131(Pt 12):3266–3276.

45. Sabri O, Sabbagh MN, Seibyl J, Barthel H, Akatsu H, Ouchi Y, et al. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer's disease: phase 3 study. Alzheimers Dement. 2015; 11:964–974.

46. Rusinek H, De Santi S, Frid D, Tsui WH, Tarshish CY, Convit A, et al. Regional brain atrophy rate predicts future cognitive decline: 6-year longitudinal MR imaging study of normal aging. Radiology. 2003; 229:691–696.

47. McGowan E, Pickford F, Kim J, Onstead L, Eriksen J, Yu C, et al. Abeta42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005; 47:191–199.
48. Mori E, Lee K, Yasuda M, Hashimoto M, Kazui H, Hirono N, et al. Accelerated hippocampal atrophy in Alzheimer's disease with apolipoprotein E epsilon4 allele. Ann Neurol. 2002; 51:209–214.
49. Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993; 90:1977–1981.

50. Strittmatter WJ, Saunders AM, Goedert M, Weisgraber KH, Dong LM, Jakes R, et al. Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: implications for Alzheimer disease. Proc Natl Acad Sci U S A. 1994; 91:11183–11186.

51. Sunderland T, Mirza N, Putnam KT, Linker G, Bhupali D, Durham R, et al. Cerebrospinal fluid beta-amyloid1-42 and tau in control subjects at risk for Alzheimer's disease: the effect of APOE epsilon4 allele. Biol Psychiatry. 2004; 56:670–676.

52. Walsh DM, Selkoe DJ. Deciphering the molecular basis of memory failure in Alzheimer's disease. Neuron. 2004; 44:181–193.

53. Conforti L, Adalbert R, Coleman MP. Neuronal death: where does the end begin? Trends Neurosci. 2007; 30:159–166.

54. Rhinn H, Fujita R, Qiang L, Cheng R, Lee JH, Abeliovich A. Integrative genomics identifies APOE ε4 effectors in Alzheimer's disease. Nature. 2013; 500:45–50.

55. Desikan RS, Thompson WK, Holland D, Hess CP, Brewer JB, Zetterberg H, et al. The role of clusterin in amyloid-β-associated neurodegeneration. JAMA Neurol. 2014; 71:180–187.

56. Albuquerque MS, Mahar I, Davoli MA, Chabot JG, Mechawar N, Quirion R, et al. Regional and sub-regional differences in hippocampal GABAergic neuronal vulnerability in the TgCRND8 mouse model of Alzheimer's disease. Front Aging Neurosci. 2015; 7:30.

57. Goutagny R, Krantic S. Hippocampal oscillatory activity in Alzheimer's disease: toward the identification of early biomarkers? Aging Dis. 2013; 4:134–140.

58. Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, et al. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007; 316:750–754.

59. Seward ME, Swanson E, Norambuena A, Reimann A, Cochran JN, Li R, et al. Amyloid-β signals through tau to drive ectopic neuronal cell cycle re-entry in Alzheimer's disease. J Cell Sci. 2013; 126(Pt 5):1278–1286.

60. Lane-Donovan C, Philips GT, Herz J. More than cholesterol transporters: lipoprotein receptors in CNS function and neurodegeneration. Neuron. 2014; 83:771–787.

61. Krstic D, Knuesel I. Deciphering the mechanism underlying late-onset Alzheimer disease. Nat Rev Neurol. 2013; 9:25–34.

62. Kim DH, Yeo SH, Park JM, Choi JY, Lee TH, Park SY, et al. Genetic markers for diagnosis and pathogenesis of Alzheimer's disease. Gene. 2014; 545:185–193.

63. Leduc V, Domenger D, De Beaumont L, Lalonde D, Bélanger-Jasmin S, Poirier J. Function and comorbidities of apolipoprotein e in Alzheimer's disease. Int J Alzheimers Dis. 2011; 2011:974361.
64. D'Onofrio G, Sancarlo D, Panza F, Copetti M, Cascavilla L, Paris F, et al. Neuropsychiatric symptoms and functional status in Alzheimer's disease and vascular dementia patients. Curr Alzheimer Res. 2012; 9:759–771.
65. Chen CH, Mizuno T, Elston R, Kariuki MM, Hall K, Unverzagt F, et al. A comparative study to screen dementia and APOE genotypes in an ageing East African population. Neurobiol Aging. 2010; 31:732–740.

66. Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, et al. Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science. 2010; 330:1774.

67. Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011; 3:89ra57.

68. Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998; 279:751–755.

69. Hendrie HC, Murrell J, Gao S, Unverzagt FW, Ogunniyi A, Hall KS. International studies in dementia with particular emphasis on populations of African origin. Alzheimer Dis Assoc Disord. 2006; 20:3 Suppl2. S42–S46.

70. Evans DA, Bennett DA, Wilson RS, Bienias JL, Morris MC, Scherr PA, et al. Incidence of Alzheimer disease in a biracial urban community: relation to apolipoprotein E allele status. Arch Neurol. 2003; 60:185–189.

71. Holtzman DM. CSF biomarkers for Alzheimer's disease: current utility and potential future use. Neurobiol Aging. 2011; 32:Suppl 1. S4–S9.

72. Alzheimer A. Die Seelenstorungen auf arteriscleroticsher Grundlage. Allgem Z Psychiatr Psych Gerichtl Med. 1902; 59:695–711.

73. Weggen S, Beher D. Molecular consequences of amyloid precursor protein and presenilin mutations causing autosomal-dominant Alzheimer's disease. Alzheimers Res Ther. 2012; 4:9.

74. Cataldo AM, Barnett JL, Pieroni C, Nixon RA. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer's disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci. 1997; 17:6142–6151.

75. Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer's disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000; 157:277–286.

76. Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengârd JH, et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000; 67:881–900.

77. McIntosh AM, Bennett C, Dickson D, Anestis SF, Watts DP, Webster TH, et al. The apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes). PLoS One. 2012; 7:e47760.

78. Finch CE, Stanford CB. Meat-adaptive genes and the evolution of slower aging in humans. Q Rev Biol. 2004; 79:3–50.

79. Eisenberg DT, Kuzawa CW, Hayes MG. Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. Am J Phys Anthropol. 2010; 143:100–111.
80. Kwon OD, Choi SY, Park JH, Yoon CH, Kwon HH, Shin IH. Association of apolipoprotein E gene polymorphism with cognitive function of the elderly residents in a rural community. J Korean Neurol Assoc. 2009; 27:362–368.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download