Abstract
Background and Purpose
Amyloid beta (Aβ) is the main component of amyloid plaques, which are deposited in the brains of patients with Alzheimer's disease (AD). Biochemical and animal studies support the central role of Aβ in AD pathogenesis. Despite several investigations focused on the pathogenic mechanisms of Aβ, it is still unclear how Aβ accumulates in the central nervous system and subsequently initiates the disease at the cellular level. In this study, we investigated the pathogenic mechanisms of Aβ using proteomics and antibody microarrays.
Methods
To evaluate the effect of Aβ on neural stem cells (NSCs), we treated primary cultured cortical NSCs with several doses of Aβ for 48 h. A 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay, trypan blue staining, and bromodeoxyuridine cell proliferation assay were performed. We detected several intracellular proteins that may be associated with Aβ by proteomics and Western blotting analysis.
Results
Various viability tests showed that Aβ decreased NSCs viability and cell proliferation in a concentration-dependent manner. Aβ treatment significantly decreased lactate dehydrogenase B, high-mobility group box 1, aldolase C, Ezrin, and survival signals including phosphorylated phosphoinositide 3-kinase, Akt, and glycogen synthase kinase-3β.
Alzheimer's disease (AD) is a progressive disease that is characterized by impaired memory and other important cognitive functions. It is the primary cause of dementia worldwide.1 Advanced age is considered the most important risk factor for the disease. Thus, the increased lifespan expectancy would rapidly raise the number of patients diagnosed with AD.2 Therefore several studies have investigated the pathophysiology of AD in the past decades. Further, several factors have been associated with disease progression and a few hypotheses have been proposed to explain the initiation and progression of the neurodegenerative process in AD.3 The “amyloid hypothesis,” states that amyloid beta (Aβ) plays a major role in the origin and progression of neurodegenerative changes seen in these patients.45 Aβ is the main component of amyloid plaques, which are deposited in the brains of patients with AD, and many biochemical and animal studies support the central role of Aβ in AD pathogenesis.67 These studies found that Aβ toxicity is mediated by several mechanisms including oxidative stress, mitochondrial diffusion, alterations in membrane permeability, inflammation, synaptic dysfunction, and excitotoxicity via interaction with neurotransmitter receptors.8910 Despite a plethora of efforts focused on the pathogenic mechanisms of Aβ, it is still unclear how Aβ accumulates in the central nervous system to trigger the disease at the cellular level.
The emergence of proteomics, the large-scale analysis of proteins, has tremendously influenced the study of cellular mechanisms.1112 The study of proteins can facilitate the analysis of protein function in cells. Proteomics is enabled by the accumulation of both DNA and protein sequence databases, improvements in mass spectrometry, and the development of computer algorithms for database search. Proteomics provides significant biological insights into specific protein-protein interactions, evaluation of proteins in subcellular compartments and elucidate protein biochemistry and the underlying cellular mechanisms. In this study, we investigated the factors associated with the pathogenic mechanisms of Aβ using proteomics.
Dulbecco's modified Eagle's medium (DMEM, high glucose) was purchased from GIBCO (Invitrogen Corporation, Grand Island, NY, USA). Amyloid-β(25-35) (Aβ25-35), protein protease inhibitor cocktail, trypan blue solution, insulin, and DNase I were obtained from Sigma-Aldrich (St. Louis, MO, USA).
All procedures using animals were consistent with Hanyang University's guidelines for the care and use of laboratory animals. We made every effort to minimize the number of animals used and animal suffering. Each animal was utilized only once.
Embryonic brain tissue was dissected from the cortex, lateral ganglionic eminence (anlage of striatum), and ventral midbrain in embryonic day 12–14 (E12–E14) rats (Sprague-Dawley, KOATECK, Seoul, Korea).13 After mechanical trituration, 20000 cells per cm2 were plated in culture dishes precoated with poly-L-ornithine (PO)/fibronectin (FN) in N2B medium [DMEM/F12, 4.4 µM insulin, 100 mg/L transferrin, 30 nM selenite, 0.6 µM putrescine, 20 nM progesterone, 0.2 mM ascorbic acid, 2 mM L-glutamine, 8.6 mM D(+) glucose, 20 mM NaHCO3, B27 (Invitrogen, Carlsbad, CA, USA)]. Following supplementation with basic fibroblast growth factor (bFGF, 20 ng/mL, R&D Systems, Minneapolis, MN, USA) and epidermal growth factor (EGF, 20 ng/mL, R&D Systems) they were cultured for 4–6 days as a monolayer on the adherent surface. To obtain a uniform population of neural stem cells (NSCs), clusters of cells formed by the proliferation of NSCs with mitogens (bFGF and EGF) were passaged by dissociating them into single cells, and plating them onto freshly PO/FN-coated coverslips (12-mm diameter; Marienfeld GmbH & Co. KG, Lauda-Knlgshofen, Germany).14 All data in this study were obtained from passaged cultures grown on the adherent surface. Cultures were maintained at 37℃ in a 5% CO2 incubator. Media were changed every other day, and mitogens were added daily.
To measure the effects of Aβ25-35 oligomers on NSC proliferation, NSCs were simultaneously treated with several concentrations of Aβ25-35 (0, 5, 10, 20, or 40 µM) for 48 h. Soluble oligomeric forms of Aβ25-35 were prepared as reported by Dahlgren et al.15 Briefly, Aβ25-35 was dissolved at a concentration of 1 mM in hexafluoroisopropanol (Sigma) and separated into aliquots in sterile microcentrifuge tubes. Hexafluoroisopropanol was removed under vacuum in a Speed-Vac, and the peptide film was stored desiccated at −20℃. To prepare oligomers, the peptide was first suspended in dry dimethyl sulfoxide (Me2SO, Sigma) at a concentration of 5 mM, and Ham's F-12 (phenol red-free, BioSource, Camarillo, CA, USA) was added to bring the final concentration to 1 mM. This was followed by 24-h incubation at 4℃. Plates were washed carefully more than three times with phosphate-buffered saline (PBS), and cell viability was measured with 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) and trypan blue assays.
MTT is absorbed into cells and transformed into formazan by mitochondrial succinate dehydrogenase. Accumulation of formazan directly reflects mitochondrial activity, which is an indirect measure of cell viability. Cells were plated in 96-well plates at a density of 1×104 cells/well in 200 µL of culture medium, and 50 µL of 2 mg/mL MTT (Sigma) was added to each well. An aliquot (220 µL) of the resulting solution was removed from each well, followed by the addition of 150 µL dimethyl sulfoxide. The precipitate from each well was resuspended on a microplate mixer for 10 min, and optical densities (OD) at 540 nm were measured using a plate reader. All results were normalized to OD values measured in an identically treated well without cultured cells. For trypan blue staining, 10 µL of dissociated cells from each sample were incubated with 10 µL of trypan blue solution (BMS, Seoul, Korea) for 2 min. Unstained live cells were counted on a hemocytometer.16
After 48 h of treatment, cells were incubated in bromodeoxyuridine (BrdU)-labeling medium (10 µM BrdU) for 1 h, and the cell proliferation assay was performed using the BrdU Labeling and Detection Kit (Roche Boehringer-Mannheim, IN, USA), according to the manufacturer's instructions. The proportion of stained cells in the 1000 cells was counted under a light microscope at 400× magnification.
We conducted proteomics analyses as reported by our previous study.17 Cultured NSC pellets were washed twice with ice-cold PBS and sonicated for 10 s with a Sonoplus (Bandelin Electronics, Berlin, Germany) in a sample lysis solution composed of 7 M urea and 2 M thiourea with 4% (w/v) 3-[(3-cholamidopropyl) dimethy-lammonio]-1-propanesulfonate (CHAPS), 1% (w/v) dithiothreitol (DTT), 2% (v/v) pharmalyte, and 1 mM benzamidine. Proteins were extracted for 1 h at room temperature by vortexing. After centrifugation at 15000 g for 1 h at 15℃, the insoluble material was discarded and the soluble fraction was used for two-dimensional (2D) gel electrophoresis. Protein concentrations were determined using the Bradford method.
Dry immobilized pH gradient strips (4–10 NL IPG, 24 cm, Genomine, Pohang, Korea) were equilibrated for 12–16 h with 7 M urea and 2 M thiourea containing 2% CHAPS, 1% DTT, and 1% pharmalyte and loaded with 200 mg of sample. Isoelectric focusing (IEF) was performed at 20℃ using a Multiphor II electrophoresis unit and EPS 3500 XL power supply (Amersham Biosciences, Buckinghamshire, UK) according to the manufacturer's instructions.
IEF was conducted by linearly increasing the voltage from 150 V to 3500 V over 3 h for sample entry followed by maintenance at a constant 3500 V. IEF was complete after 96 kVh. Prior to running the second dimension, the strips were incubated for 10 min in equilibration buffer [50 mM Tris-Cl, pH 6.8, containing 6 M urea, 2% sodium dodecyl sulfate (SDS), and 30% glycerol], first with 1% DTT and then with 2.5% iodoacetamide. The equilibrated strips were then inserted onto SDS polyacrylamide gel electrophoresis (SDS-PAGE) gels (20×24 cm, 10–16%). SDS-PAGE was performed using the Hoefer DALT 2D system (Amersham Biosciences) according to the manufacturer's instructions. The 2D gels were run at 20℃ for 1700 Vh.
Quantitative analyses of digitized images were performed using PDQuest (version 7.0, Bio-Rad, Hercules, CA, USA) software according to the manufacturer's protocols. The intensity of each spot was normalized to the total valid spot intensity. Protein spots with at least two-fold difference in expression compared with the control or normal samples were selected. For protein identification by peptide mass fingerprinting, protein spots were excised, digested with trypsin (Promega, Madison, WI, USA), mixed with a-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% trifluoroacetic acid and subjected to matrix-assisted laser desorption/ionization-time of flight analysis (Microflex LRF 20, Bruker Daltonics, Billerica, MA, USA). Spectra were collected from 300 shots per spectrum over the m/z range of 600–3000 and calibrated by a two-point internal calibration using trypsin auto-digestion peaks (m/z 842.5099 and 2211.1046). The peak list was generated using Flex Analysis 3.0 (Bruker Daltonics). The thresholds used for peak selection were as follows: 500 for a minimum resolution of monoisotopic mass and 5 for signal-to-noise. The search program MASCOT, developed by Matrix Science (http://www.matrixscience.com/), was used for protein identification by peptide mass fingerprinting. The following parameters were used for the database search: trypsin as the cleaving enzyme, a maximum of one missed cleavage, iodoacetamide (Cys) as a complete modification, oxidation (Met) as a partial modification, monoisotopic mass, and a mass, tolerance of 0.1 Da. The peptide mass fingerprinting acceptance criteria were based on probability scoring.
The levels of p85α phosphoinositide 3-kinase (PI3K), phosphorylated Akt (pAkt) (Ser 473), and phosphorylated glycogen synthase kinase-3β (pGSK-3β) (Ser9) were analyzed by Western blot. Briefly, 5×106 cells were washed twice in cold PBS and incubated in lysis buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 0.02% sodium azide, 0.2% SDS, 100 µg/mL phenylmethylsulfonylfluoride, 50 µL/mL aprotinin, 1% Igepal 630, 100 mM NaF, 0.5% sodium deoxycholate, 0.5 mM EDTA, and 0.1 mM EGTA] for 10 min on ice. Cell lysates were centrifuged at 10000 µg and evaluated for levels of p85α PI3K, pAkt, and pGSK-3β. Protein concentrations of cell lysates and postmitochondrial fractions were determined using a Bio-Rad protein assay kit (Bio-Rad). Samples containing equal amounts (20 µg) of protein were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes (Amersham Pharmacia Biotech, Buckinghamshire, UK). Membranes were blocked using 5% skim milk before incubation with specific primary antibodies against p85α PI3K (1:1000, Sigma), pAkt (1:500, Cell Signaling, Beverly, MA, USA), and pGSK-3β (Ser9) (1:1000, Santa Cruz Biotech, Santa Cruz, CA, USA). Membranes were washed with Tris-buffered saline containing 0.05% Tween-20 and processed using an HRP-conjugated anti-rabbit or anti-mouse antibody (Amersham Pharmacia Biotech, Piscataway, NJ, USA) followed by ECL detection (Amersham Pharmacia Biotech).18 The results from Western blots were quantified using an image analyzer (Quantity One-4,2,0, Bio-Rad).
All data are presented as the mean±standard deviation from five independent experiments. Statistical comparisons of viability among the different treatment groups were performed with Tukey's test after a one-way analysis of variance. p-values less than 0.05 were considered statistically significant.
To evaluate the effect of Aβ25-35 on viability, NSCs were treated with different concentrations of Aβ25-35 (0, 5, 10, 20, or 40 µM) for 48 h. Cell viability was measured with the MTT assay, and surviving cells were counted with trypan blue staining. The viability of NSCs was not significantly decreased by treatment with 5 µM Aβ25-35, but the viability of cells treated with concentrations greater than 10 µM was significantly decreased in a concentration-dependent manner (Fig. 1).
BrdU labeling showed that the proliferation of NSCs treated with Aβ25-35 at concentrations greater than 5 µM was significantly decreased in a concentration-dependent manner (Fig. 2). Based on these results, 20 µM Aβ25-35 was selected as an optimal Aβ25-35 concentration for subsequent experiments based on 60–70% cell viability.
Proteomics studies were performed to assess the effect of 20 µM Aβ25-35 on the synthesis of intracellular proteins in NSCs. We found that the expression of several intracellular proteins was altered by 20 µM Aβ25-35. Theoretical and experimental molecular weights (MWs) as well as isoelectric points (pIs) calculated from 2D-E maps are shown in Fig. 3. The values of MW and pIs of each protein corresponded roughly to its position on the 2D-E gel. Deviations from theoretical MW and pI were found to be substantially lower than in several cases. Specifically, the expressions of LDHB, HMGB1, ALDOC, and Ezrin were reduced by exposure to 20 µM Aβ25-35 (Fig. 3). Using the Western blot, we ascertained the effects of 20 µM Aβ25-35 oligomer on the intracellular signaling of proteins, which play a critical role in NSC proliferation. We determined the expression of p85α PI3K, pAkt (Ser473), and pGSK-3β (Ser9) in the PI3K pathway that plays a pivotal role in the proliferation of NSCs. Compared with untreated cells (100±10, 100±11, 100±9, and 100±8%, respectively), the immunoreactivities (IRs) of p85α PI3K, pAkt (Ser473), and pGSK-3α (Ser9) were significantly decreased in 20 µM Aβ25-35 oligomer-treated cells (43±8, 47±7 and 72±10%, respectively, p<0.05) (Fig. 4).
In the present study, we investigated the effects of Aβ25-35 on NSCs and the intracellular protein levels of NSCs in the pathogenesis of Aβ25-35 oligomers. Using proteomics, we confirmed the reduced expression of lactate dehydrogenase B (LDHB), high-mobility group box 1 (HMGB1), aldolase C (ALDOC), and Ezrin following treatment with Aβ25-35 oligomer. LDHB is a subunit of the lactate dehydrogenase enzyme,19 which is found throughout the body, and plays an important role in cellular energy production. In the final step of glucose breakdown, most forms of lactate dehydrogenase enzyme convert pyruvate into lactate, which is used by the body for energy.20 Previous investigations revealed that LDHB mediated the resistance to Aβ toxicity in the neuronal cells by reducing oxidative stress.21 HMGB1 plays a key role in inflammation, and immunostimulation and chemotactic processes.222324 Increased levels of HMGB1 have been detected in several neurodegenerative diseases. HMGB1 is a risk factor for memory impairment, chronic neurodegeneration, and progression of neuro-inflammation in AD and it has a neuroprotective role in Huntington's disease.2526 In this study, treatment of Aβ oligomer decreased the expression of HMGB1. Thus, HMGB1 plays a key dual role in neurodegeneration, although the underlying pathological mechanisms remain uncertain. ALDOC is an important enzyme in glycolysis, and is predominantly expressed in neuronal tissue specifically in the hippocampus and Purkinje cells of the brain.2728 In previous studies, the ALDOC level was decreased in AD brain,21 however, the precise ALDOC-mediated mechanisms remain unknown. Ezrin, one of the ezrin-radixin-moesin proteins, is involved in cell-surface phenomena and is expressed in a variety of tissues.29 Ezrin typically concentrates at the apical surface of polarized epithelia and its upregulation enhances the metastatic potential in various tumor types.3031 In Parkinson's disease, the expression of Ezrin specifically inhibits α-synuclein fibrillization and toxicity.323334 Further investigation into the role of Ezrin in the pathophysiology of Alzheimer's disease is needed.
Our Western blotting results showed that the expression of survival-related proteins such as p85α PI3K, pAkt (Ser473), and pGSK-3β (Ser9) was also reduced by Aβ25-35. The expression of these proteins in PI3K pathway is essential for the survival and self-renewal of NSCs,35 and activation of this pathway is important for maintaining the pluripotency of NSCs.36 In the first step of PI3K pathway, activated PI3K phosphorylates its downstream target, Akt, and GSK-3β. The pAkt also inhibits GSK-3β by phosphorylating it at Ser932. While the active form of GSK-3β inhibits HSTF-1 and activates the mitochondrial death pathway,3536 inactivation of the pGSK-3β by pAkt is important functionally in a variety of stem cell types.37 In previous studies, we confirmed that Aβ toxicity in neuronal cells or NSCs, and the protective effects of neuroprotectants such as coenzyme Q10, were mediated via PI3K pathway.3839
The study limitations are as follows. First, this study was performed under in vitro conditions, which preclude any application to in vivo conditions. Second, no mechanistic study was conducted to confirm the targets of intracellular proteins, which were influenced by Aβ25-35. The precise mechanism of action should be delineated in further studies of Aβ25-35 based on our results. Finally, AD pathogenesis is associated with adult hippocampal NSCs, whereas only embryonic cortical NSCs were available for use in this in vitro study. To address this limitation, an in vitro mouse model of AD is needed.
Taken together, these results suggest that several intracellular proteins detected by proteomics and Western blotting analyses hold the clues to Aβ pathogenesis. Further studies should be conducted with a focus on these intracellular proteins.
Figures and Tables
 | Fig. 1Viability of NSCs injured by Aβ25-35. MTT assay and TBS show that the viability of NSCs was decreased in a concentration-dependent manner following treatment with more than 10 µM Aβ25-35 oligomer. *p<0.05 (vs. control group). Aβ: amyloid beta, con: control group, MTT: 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide, NSC: neural stem cell, TBS: trypan blue staining. |
 | Fig. 2Proliferation of NSCs injured by Aβ25-35. BrdU assay showed that the proliferation of NSCs was decreased in a concentration-dependent manner when the cells were treated with more than 10 µM Aβ25-35 oligomer. *p<0.05 (vs. control group). Aβ: amyloid beta, BrdU: bromodeoxyuridine, con: control group, NSC: neural stem cell. |
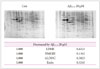 | Fig. 3Effects of Aβ25-35 on intracellular protein levels of NSCs in proteomics. Proteomics showed the effects of 20 µM Aβ25-35 on various intracellular proteins in NSCs. Representative 2D-E image obtained from control group and following the treatment of 20 µM Aβ25-35 oligomer group. Protein spots with at least a two-fold difference in expression compared with the control or normal samples were selected. The proteomics results confirmed that 20 µM Aβ25-35 decreased the expression of proteins associated with cell survival. Aβ: amyloid beta, ALDOC: aldolase C, con: control group, HMGB1: high-mobility group box 1, LDHB: lactate dehydrogenase B, NSC: neural stem cell. |
 | Fig. 4Effects of Aβ25-35 on intracellular protein levels of NSCs in Western blot. Western blotting analysis showed that treatment with 20 µM Aβ25-35 significantly inhibited the expression of proliferation-related intracellular signaling proteins of NSCs. *p<0.05 (vs. control group). Aβ: amyloid beta, NSC: neural stem cell. |
Acknowledgements
This work was supported by the Basic Science Research Program of the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (2015R1A2A2A04004865), the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), the Ministry of Health & Welfare, Republic of Korea (grant number: HI17C2160), the Korea Drug Development Fund (KDDF) funded by the Ministry of Science and ICT, Ministry of Trade, Industry & Energy, and Ministry of Health & Welfare (KDDF-201609-02, Republic of Korea), and by the Medical Research Center (2017R1A5A2015395).
References
1. Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, et al. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005; 366:2112–2117.

4. Lewczuk P, Mroczko B, Fagan A, Kornhuber J. Biomarkers of Alzheimer's disease and mild cognitive impairment: a current perspective. Adv Med Sci. 2015; 60:76–82.

5. Nhan HS, Chiang K, Koo EH. The multifaceted nature of amyloid precursor protein and its proteolytic fragments: friends and foes. Acta Neuropathol. 2015; 129:1–19.

6. Vetrivel KS, Thinakaran G. Amyloidogenic processing of beta-amyloid precursor protein in intracellular compartments. Neurology. 2006; 66:2 Suppl 1. S69–S73.
7. Atwood CS, Obrenovich ME, Liu T, Chan H, Perry G, Smith MA, et al. Amyloid-beta: a chameleon walking in two worlds: a review of the trophic and toxic properties of amyloid-beta. Brain Res Brain Res Rev. 2003; 43:1–16.

8. Muresan V, Ladescu Muresan Z. Amyloid-β precursor protein: multiple fragments, numerous transport routes and mechanisms. Exp Cell Res. 2015; 334:45–53.

9. Cappai R. Making sense of the amyloid precursor protein: its tail tells an interesting tale. J Neurochem. 2014; 130:325–327.

10. Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008; 283:29615–29619.

11. Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, et al. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003; 3:36–44.

12. Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002; 1:845–867.

13. Park CH, Kang JS, Yoon EH, Shim JW, Suh-Kim H, Lee SH. Proneural bHLH neurogenin 2 differentially regulates Nurr1-induced dopamine neuron differentiation in rat and mouse neural precursor cells in vitro. FEBS Lett. 2008; 582:537–542.

14. Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996; 10:3129–3140.

15. Dahlgren KN, Manelli AM, Stine WB Jr, Baker LK, Krafft GA, LaDu MJ. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J Biol Chem. 2002; 277:32046–32053.

16. Noh MY, Koh SH, Kim Y, Kim HY, Cho GW, Kim SH. Neuroprotective effects of donepezil through inhibition of GSK-3 activity in amyloid-beta-induced neuronal cell death. J Neurochem. 2009; 108:1116–1125.

17. Park HH, Yu HJ, Kim S, Kim G, Choi NY, Lee EH, et al. Neural stem cells injured by oxidative stress can be rejuvenated by GV1001, a novel peptide, through scavenging free radicals and enhancing survival signals. Neurotoxicology. 2016; 55:131–141.

18. Lee KY, Koh SH, Noh MY, Kim SH, Lee YJ. Phosphatidylinositol-3-kinase activation blocks amyloid beta-induced neurotoxicity. Toxicology. 2008; 243:43–50.

19. Adeva-Andany M, López-Ojén M, Funcasta-Calderón R, Ameneiros-Rodríguez E, Donapetry-García C, Vila-Altesor M, et al. Comprehensive review on lactate metabolism in human health. Mitochondrion. 2014; 17:76–100.

20. Pineda JR, Callender R, Schwartz SD. Ligand binding and protein dynamics in lactate dehydrogenase. Biophys J. 2007; 93:1474–1483.

21. Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004; 101:2173–2178.

22. Fang P, Schachner M, Shen YQ. HMGB1 in development and diseases of the central nervous system. Mol Neurobiol. 2012; 45:499–506.

23. Tang SC, Arumugam TV, Xu X, Cheng A, Mughal MR, Jo DG, et al. Pivotal role for neuronal Toll-like receptors in ischemic brain injury and functional deficits. Proc Natl Acad Sci U S A. 2007; 104:13798–13803.

24. Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002; 418:191–195.

25. Tahara K, Kim HD, Jin JJ, Maxwell JA, Li L, Fukuchi K. Role of toll-like receptor signalling in Abeta uptake and clearance. Brain. 2006; 129(Pt 11):3006–3019.

26. Fujita K, Motoki K, Tagawa K, Chen X, Hama H, Nakajima K, et al. HMGB1, a pathogenic molecule that induces neurite degeneration via TLR4-MARCKS, is a potential therapeutic target for Alzheimer's disease. Sci Rep. 2016; 6:31895.

27. Arakaki TL, Pezza JA, Cronin MA, Hopkins CE, Zimmer DB, Tolan DR, et al. Structure of human brain fructose 1,6-(bis)phosphate aldolase: linking isozyme structure with function. Protein Sci. 2004; 13:3077–3084.

28. Sekar Y, Moon TC, Slupsky CM, Befus AD. Protein tyrosine nitration of aldolase in mast cells: a plausible pathway in nitric oxide-mediated regulation of mast cell function. J Immunol. 2010; 185:578–587.

29. Osawa H, Smith CA, Ra YS, Kongkham P, Rutka JT. The role of the membrane cytoskeleton cross-linker ezrin in medulloblastoma cells. Neuro Oncol. 2009; 11:381–393.

30. Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993; 105(Pt 4):1025–1043.

31. Bretscher A. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr Opin Cell Biol. 1999; 11:109–116.

32. Li Y, Harada T, Juang YT, Kyttaris VC, Wang Y, Zidanic M, et al. Phosphorylated ERM is responsible for increased T cell polarization, adhesion, and migration in patients with systemic lupus erythematosus. J Immunol. 2007; 178:1938–1947.

33. Johnson MW, Miyata H, Vinters HV. Ezrin and moesin expression within the developing human cerebrum and tuberous sclerosis-associated cortical tubers. Acta Neuropathol. 2002; 104:188–196.

34. Yamada M, Iwatsubo T, Mizuno Y, Mochizuki H. Overexpression of alpha-synuclein in rat substantia nigra results in loss of dopaminergic neurons, phosphorylation of alpha-synuclein and activation of caspase-9: resemblance to pathogenetic changes in Parkinson's disease. J Neurochem. 2004; 91:451–461.

35. Pap M, Cooper GM. Role of translation initiation factor 2B in control of cell survival by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3beta signaling pathway. Mol Cell Biol. 2002; 22:578–586.

36. Frame S, Cohen P, Biondi RM. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell. 2001; 7:1321–1327.

37. Liu S, Liu S, Wang X, Zhou J, Cao Y, Wang F, et al. The PI3K-Akt pathway inhibits senescence and promotes self-renewal of human skin-derived precursors in vitro. Aging Cell. 2011; 10:661–674.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download